A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration
- PMID: 20157114
- PMCID: PMC2857069
- DOI: 10.1074/jbc.M109.045161
A structural determinant that renders G alpha(i) sensitive to activation by GIV/girdin is required to promote cell migration
Abstract
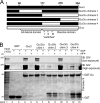
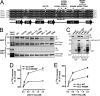
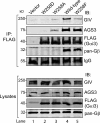
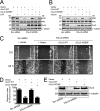
Although several non-receptor activators of heterotrimeric G proteins have been identified, the structural features of G proteins that determine their interaction with such activators and the subsequent biological effects are poorly understood. Here we investigated the structural determinants in G alpha(i3) necessary for its regulation by GIV/girdin, a guanine-nucleotide exchange factor (GEF) that activates G alpha(i) subunits. Using G protein activity and in vitro pulldown assays we demonstrate that G alpha(i3) is a better substrate for GIV than the highly homologous G alpha(o). We identified Trp-258 in the G alpha(i) subunit as a novel structural determinant for GIV binding by comparing GIV binding to G alpha(i3)/G alpha(o) chimeras. Mutation of Trp-258 to the corresponding Phe in G alpha(o) decreased GIV binding in vitro and in cultured cells but did not perturb interaction with other G alpha-binding partners, i.e. G betagamma, AGS3 (a guanine nucleotide dissociation inhibitor), GAIP/RGS19 (a GTPase-activating protein), and LPAR1 (a G protein-coupled receptor). Activation of G alpha(i3) by GIV was also dramatically reduced when Trp-258 was replaced with Tyr, Leu, Ser, His, Asp, or Ala, highlighting that Trp is required for maximal activation. Moreover, when mutant G alpha(i3) W258F was expressed in HeLa cells they failed to undergo cell migration and to enhance Akt signaling after growth factor or G protein-coupled receptor stimulation. Thus activation of G alpha(i3) by GIV is essential for biological functions associated with G alpha(i3) activation. In conclusion, we have discovered a novel structural determinant on G alpha(i) that plays a key role in defining the selectivity and efficiency of the GEF activity of GIV on G alpha(i) and that represents an attractive target site for designing small molecules to disrupt the G alpha(i)-GIV interface for therapeutic purposes.
Figures






Similar articles
-
A GDI (AGS3) and a GEF (GIV) regulate autophagy by balancing G protein activity and growth factor signals.Mol Biol Cell. 2011 Mar 1;22(5):673-86. doi: 10.1091/mbc.E10-08-0738. Epub 2011 Jan 5. Mol Biol Cell. 2011. PMID: 21209316 Free PMC article.
-
GIV is a nonreceptor GEF for G alpha i with a unique motif that regulates Akt signaling.Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3178-83. doi: 10.1073/pnas.0900294106. Epub 2009 Feb 11. Proc Natl Acad Sci U S A. 2009. PMID: 19211784 Free PMC article.
-
GIV/Girdin activates Gαi and inhibits Gαs via the same motif.Proc Natl Acad Sci U S A. 2016 Sep 27;113(39):E5721-30. doi: 10.1073/pnas.1609502113. Epub 2016 Sep 12. Proc Natl Acad Sci U S A. 2016. PMID: 27621449 Free PMC article.
-
Structural determinants involved in the formation and activation of G protein betagamma dimers.Neurosignals. 2009;17(1):82-99. doi: 10.1159/000186692. Epub 2009 Feb 12. Neurosignals. 2009. PMID: 19212142 Free PMC article. Review.
-
GIV/Girdin transmits signals from multiple receptors by triggering trimeric G protein activation.J Biol Chem. 2015 Mar 13;290(11):6697-704. doi: 10.1074/jbc.R114.613414. Epub 2015 Jan 20. J Biol Chem. 2015. PMID: 25605737 Free PMC article. Review.
Cited by
-
Involvement of Girdin in the determination of cell polarity during cell migration.PLoS One. 2012;7(5):e36681. doi: 10.1371/journal.pone.0036681. Epub 2012 May 4. PLoS One. 2012. PMID: 22574214 Free PMC article.
-
The mechanism of Girdin in degenerative brain disease caused by high glucose stimulation.Front Endocrinol (Lausanne). 2022 Oct 3;13:892897. doi: 10.3389/fendo.2022.892897. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36329890 Free PMC article.
-
Direct interrogation of context-dependent GPCR activity with a universal biosensor platform.bioRxiv [Preprint]. 2024 Jan 2:2024.01.02.573921. doi: 10.1101/2024.01.02.573921. bioRxiv. 2024. Update in: Cell. 2024 Mar 14;187(6):1527-1546.e25. doi: 10.1016/j.cell.2024.01.028. PMID: 38260348 Free PMC article. Updated. Preprint.
-
Revealing the Activity of Trimeric G-proteins in Live Cells with a Versatile Biosensor Design.Cell. 2020 Aug 6;182(3):770-785.e16. doi: 10.1016/j.cell.2020.06.020. Epub 2020 Jul 6. Cell. 2020. PMID: 32634377 Free PMC article.
-
Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16394-16403. doi: 10.1073/pnas.1906658116. Epub 2019 Jul 30. Proc Natl Acad Sci U S A. 2019. PMID: 31363053 Free PMC article.
References
-
- De Vries L., Zheng B., Fischer T., Elenko E., Farquhar M. G. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 235–271 - PubMed
-
- Gilman A. G. (1987) Annu. Rev. Biochem. 56, 615–649 - PubMed
-
- Ross E. M., Wilkie T. M. (2000) Annu. Rev. Biochem. 69, 795–827 - PubMed
-
- Oldham W. M., Hamm H. E. (2008) Nat. Rev. Mol. Cell Biol. 9, 60–71 - PubMed
-
- Overington J. P., Al-Lazikani B., Hopkins A. L. (2006) Nat. Rev. Drug Discov. 5, 993–996 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

