New mutant phenotype data curation system in the Saccharomyces Genome Database
- PMID: 20157474
- PMCID: PMC2790299
- DOI: 10.1093/database/bap001
New mutant phenotype data curation system in the Saccharomyces Genome Database
Abstract
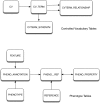
The Saccharomyces Genome Database (SGD; http://www.yeastgenome.org/) organizes and displays molecular and genetic information about the genes and proteins of baker's yeast, Saccharomyces cerevisiae. Mutant phenotype screens have been the starting point for a large proportion of yeast molecular biological studies, and are still used today to elucidate the functions of uncharacterized genes and discover new roles for previously studied genes. To greatly facilitate searching and comparison of mutant phenotypes across genes, we have devised a new controlled-vocabulary system for capturing phenotype information. Each phenotype annotation is represented as an 'observable', which is the entity, or process that is observed, and a 'qualifier' that describes the change in that entity or process in the mutant (e.g. decreased, increased, or abnormal). Additional information about the mutant, such as strain background, allele name, conditions under which the phenotype is observed, or the identity of relevant chemicals, is captured in separate fields. For each gene, a summary of the mutant phenotype information is displayed on the Locus Summary page, and the complete information is displayed in tabular format on the Phenotype Details Page. All of the information is searchable and may also be downloaded in bulk using SGD's Batch Download Tool or Download Data Files Page. In the future, phenotypes will be integrated with other curated data to allow searching across different types of functional information, such as genetic and physical interaction data and Gene Ontology annotations.Database URL:http://www.yeastgenome.org/
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

