QuickGO: a user tutorial for the web-based Gene Ontology browser
- PMID: 20157483
- PMCID: PMC2794795
- DOI: 10.1093/database/bap010
QuickGO: a user tutorial for the web-based Gene Ontology browser
Abstract
The Gene Ontology (GO) has proven to be a valuable resource for functional annotation of gene products. At well over 27 000 terms, the descriptiveness of GO has increased rapidly in line with the biological data it represents. Therefore, it is vital to be able to easily and quickly mine the functional information that has been made available through these GO terms being associated with gene products. QuickGO is a fast, web-based tool for browsing the GO and all associated GO annotations provided by the GOA group. After undergoing a redevelopment, QuickGO is now able to offer many more features beyond simple browsing. Users have responded well to the new tool and given very positive feedback about its usefulness. This tutorial will demonstrate how some of these features could be useful to the researcher wanting to discover more about their dataset, particular areas of biology or to find new ways of directing their research.Database URL:http://www.ebi.ac.uk/QuickGO.
Figures
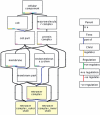
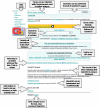











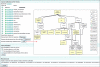




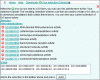
Similar articles
-
QuickGO: a web-based tool for Gene Ontology searching.Bioinformatics. 2009 Nov 15;25(22):3045-6. doi: 10.1093/bioinformatics/btp536. Epub 2009 Sep 10. Bioinformatics. 2009. PMID: 19744993 Free PMC article.
-
The GOA database in 2009--an integrated Gene Ontology Annotation resource.Nucleic Acids Res. 2009 Jan;37(Database issue):D396-403. doi: 10.1093/nar/gkn803. Epub 2008 Oct 27. Nucleic Acids Res. 2009. PMID: 18957448 Free PMC article.
-
A drug target slim: using gene ontology and gene ontology annotations to navigate protein-ligand target space in ChEMBL.J Biomed Semantics. 2016 Sep 27;7(1):59. doi: 10.1186/s13326-016-0102-0. J Biomed Semantics. 2016. PMID: 27678076 Free PMC article.
-
Gene Ontology annotation of the rice blast fungus, Magnaporthe oryzae.BMC Microbiol. 2009 Feb 19;9 Suppl 1(Suppl 1):S8. doi: 10.1186/1471-2180-9-S1-S8. BMC Microbiol. 2009. PMID: 19278556 Free PMC article. Review.
-
An Experimental Approach to Genome Annotation: This report is based on a colloquium sponsored by the American Academy of Microbiology held July 19-20, 2004, in Washington, DC.Washington (DC): American Society for Microbiology; 2004. Washington (DC): American Society for Microbiology; 2004. PMID: 33001599 Free Books & Documents. Review.
Cited by
-
A new metabolic model of Drosophila melanogaster and the integrative analysis of Parkinson's disease.Life Sci Alliance. 2023 May 26;6(8):e202201695. doi: 10.26508/lsa.202201695. Print 2023 Aug. Life Sci Alliance. 2023. PMID: 37236669 Free PMC article.
-
Cherry Blossom Forecast Based on Transcriptome of Floral Organs Approaching Blooming in the Flowering Cherry (Cerasus × yedoensis) Cultivar 'Somei-Yoshino'.Front Plant Sci. 2022 Jan 26;13:802203. doi: 10.3389/fpls.2022.802203. eCollection 2022. Front Plant Sci. 2022. PMID: 35154222 Free PMC article.
-
TGFB1-Mediated Gliosis in Multiple Sclerosis Spinal Cords Is Favored by the Regionalized Expression of HOXA5 and the Age-Dependent Decline in Androgen Receptor Ligands.Int J Mol Sci. 2019 Nov 26;20(23):5934. doi: 10.3390/ijms20235934. Int J Mol Sci. 2019. PMID: 31779094 Free PMC article.
-
Identification of 13 novel susceptibility loci for early-onset myocardial infarction, hypertension, or chronic kidney disease.Int J Mol Med. 2018 Nov;42(5):2415-2436. doi: 10.3892/ijmm.2018.3852. Epub 2018 Sep 4. Int J Mol Med. 2018. PMID: 30226566 Free PMC article.
-
Seawater salt-trapped Pseudomonas aeruginosa survives for years and gets primed for salinity tolerance.BMC Microbiol. 2019 Jun 24;19(1):142. doi: 10.1186/s12866-019-1499-2. BMC Microbiol. 2019. PMID: 31234794 Free PMC article.
References
-
- Pereira GS, Brandão RM, Giuliatti S, et al. Gene Class expression: analysis tool of Gene Ontology terms with gene expression data. Genet. Mol. Res. 2006;5:108–114. - PubMed
-
- Pirooznia M, Habib T, Perkins EJ, et al. GOfetcher: a database with complex searching facility for gene ontology. Bioinformatics. 2008;24:2561–2563. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

