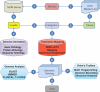
ORION-VIRCAT: a tool for mapping ICTV and NCBI taxonomies
- PMID: 20157487
- PMCID: PMC2790308
- DOI: 10.1093/database/bap014
ORION-VIRCAT: a tool for mapping ICTV and NCBI taxonomies
Abstract
Viruses, viroids and prions are the smallest infectious biological entities that depend on their host for replication. The number of pathogenic viruses is considerably large and their impact in human global health is well documented. Currently, the International Committee on the Taxonomy of Viruses (ICTV) has classified approximately 4379 virus species while the National Center for Biotechnology Information Viral Genomes Resource (NCBI-VGR) database has mapped 617 705 proteins to eight large taxonomic groups. Despite these efforts, an automated approach for mapping the ICTV master list and its officially accepted virus naming to the NCBI-VGR's taxonomical classification is not available. Due to metagenomic sequencing, it is likely that the discovery and naming of new viral species will increase by at least ten fold. Unfortunately, existing viral databases are not adequately prepared to scale, maintain and annotate automatically ultra-high throughput sequences and place this information into specific taxonomic categories. ORION-VIRCAT is a scalable and interoperable object-relational database designed to serve as a resource for the integration and verification of taxonomical classifications generated by the ICTV and NCBI-VGR. The current release (v1.0) of ORION-VIRCAT is implemented in PostgreSQL and it has been extended to ORACLE, MySQL and SyBase. ORION-VIRCAT automatically mapped and joined 617 705 entries from the NCBI-VGR to the viral naming of the ICTV. This detailed analysis revealed that 399 095 entries from the NCBI-VGR can be mapped to the ICTV classification and that one Order, 10 families, 35 genera and 503 species listed in the ICTV disagree with the the NCBI-VGR classification schema. Nevertheless, we were eable to correct several discrepancies mapping 234 000 additional entries.Database URL:http://www.orionbiosciences.com/research/orion-vircat.html.
Figures
Similar articles
-
The Genome-Based Virus Taxonomy with ICTV Database Extension.Genomics Inform. 2018 Dec;16(4):e22. doi: 10.5808/GI.2018.16.4.e22. Epub 2018 Dec 28. Genomics Inform. 2018. PMID: 30602083 Free PMC article.
-
Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV).Nucleic Acids Res. 2018 Jan 4;46(D1):D708-D717. doi: 10.1093/nar/gkx932. Nucleic Acids Res. 2018. PMID: 29040670 Free PMC article.
-
Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021).Arch Virol. 2021 Sep;166(9):2633-2648. doi: 10.1007/s00705-021-05156-1. Arch Virol. 2021. PMID: 34231026
-
Solving the species problem in viral taxonomy: recommendations on non-Latinized binomial species names and on abandoning attempts to assign metagenomic viral sequences to species taxa.Arch Virol. 2019 Sep;164(9):2223-2229. doi: 10.1007/s00705-019-04320-y. Epub 2019 Jun 18. Arch Virol. 2019. PMID: 31209597 Review.
-
ICTV Virus Taxonomy Profile: Solinviviridae.J Gen Virol. 2019 May;100(5):736-737. doi: 10.1099/jgv.0.001242. Epub 2019 Mar 5. J Gen Virol. 2019. PMID: 30835197 Review.
Cited by
-
Biosurveillance enterprise for operational awareness, a genomic-based approach for tracking pathogen virulence.Virulence. 2013 Nov 15;4(8):745-51. doi: 10.4161/viru.26893. Epub 2013 Oct 23. Virulence. 2013. PMID: 24152965 Free PMC article. Review.
-
Molecular evolution of epizootic hemorrhagic disease viruses in North America based on historical isolates using motif fingerprints.Virus Genes. 2016 Aug;52(4):495-508. doi: 10.1007/s11262-016-1332-z. Epub 2016 Apr 23. Virus Genes. 2016. PMID: 27107856
-
Enhanced taxonomy annotation of antiviral activity data from ChEMBL.Database (Oxford). 2019 Jan 1;2019:bay139. doi: 10.1093/database/bay139. Database (Oxford). 2019. PMID: 30753475 Free PMC article.
References
LinkOut - more resources
Full Text Sources