WRN helicase defective in the premature aging disorder Werner syndrome genetically interacts with topoisomerase 3 and restores the top3 slow growth phenotype of sgs1 top3
- PMID: 20157511
- PMCID: PMC2806000
- DOI: 10.18632/aging.100020
WRN helicase defective in the premature aging disorder Werner syndrome genetically interacts with topoisomerase 3 and restores the top3 slow growth phenotype of sgs1 top3
Abstract
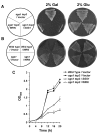
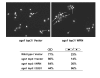
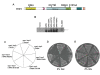
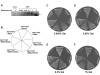
Werner syndrome (WS) is a premature aging disorder characterized by genomic instability. The WRN gene defective in WS encodes a protein with both helicase and exonuclease activities that interacts with proteins implicated in DNA metabolism. To understand its genetic functions, we examined the ability of human WRN to rescue phenotypes associated with sgs1, the sole RecQ helicase in Saccharomyces cerevisiae. WRN failed to rescue sgs1 sensitivity to the DNA damaging agent methylmethane sulfonate or replication inhibitor hydroxyurea, suggesting divergent functions of human and yeast RecQ helicases. However, physiological expression of WRN in sgs1 top3 restored top3 slow growth phenotype, whereas no effect on growth was observed with wild-type or sgs1 strains. Slow growth of WRN-transformed sgs1 top3 correlated with an elevated population of large-budded cells with undivided nuclei, indicating restoration of cell cycle delay in late S/G2 characteristic of top3. WRN helicase but not exonuclease activity was genetically required for restoration of top3 growth phenotype, demonstrating separation of function of WRN catalytic activities. A naturally occurring missense polymorphism in WRN that interferes with helicase activity abolished its ability to restore top3 slow growth phenotype. Proposed roles of WRN in genetic pathways important for the suppression of genomic instability are discussed.
Keywords: Bloom's syndrome; RecQ; Sgs1; Werner syndrome; genomic instability; helicase; topoisomerase.
Conflict of interest statement
The authors in this manuscript have no conflict of interests to declare.
Figures


Similar articles
-
Premature aging syndrome gene WRN genetically interacts with a topoisomerase.Cell Cycle. 2009 Jul 15;8(14):2143. doi: 10.4161/cc.8.14.9118. Epub 2009 Jul 15. Cell Cycle. 2009. PMID: 19587535 No abstract available.
-
SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination.Nat Genet. 2001 Jan;27(1):113-6. doi: 10.1038/83673. Nat Genet. 2001. PMID: 11138010
-
Bloom's and Werner's syndrome genes suppress hyperrecombination in yeast sgs1 mutant: implication for genomic instability in human diseases.Proc Natl Acad Sci U S A. 1998 Jul 21;95(15):8733-8. doi: 10.1073/pnas.95.15.8733. Proc Natl Acad Sci U S A. 1998. PMID: 9671747 Free PMC article.
-
[Functional analysis of yeast homologue gene associated with human DNA helicase causative syndromes].Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002;(120):53-74. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 2002. PMID: 12638184 Review. Japanese.
-
Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication.J Biochem. 2001 Apr;129(4):501-7. doi: 10.1093/oxfordjournals.jbchem.a002883. J Biochem. 2001. PMID: 11275547 Review.
Cited by
-
Genetic mutants illuminate the roles of RecQ helicases in recombinational repair or response to replicational stress.Cell Cycle. 2010 Aug 15;9(16):3139-41. doi: 10.4161/cc.9.16.12953. Epub 2010 Aug 9. Cell Cycle. 2010. PMID: 20703073 Free PMC article. No abstract available.
-
Impact papers on aging in 2009.Aging (Albany NY). 2010 Mar;2(3):111-21. doi: 10.18632/aging.100132. Epub 2010 Mar 23. Aging (Albany NY). 2010. PMID: 20351400 Free PMC article. Review.
-
Genetic studies of human DNA repair proteins using yeast as a model system.J Vis Exp. 2010 Mar 18;(37):1639. doi: 10.3791/1639. J Vis Exp. 2010. PMID: 20300059 Free PMC article.
-
Elucidation of DNA Repair Function of PfBlm and Potentiation of Artemisinin Action by a Small-Molecule Inhibitor of RecQ Helicase.mSphere. 2020 Nov 25;5(6):e00956-20. doi: 10.1128/mSphere.00956-20. mSphere. 2020. PMID: 33239368 Free PMC article.
-
Functional deficit associated with a missense Werner syndrome mutation.DNA Repair (Amst). 2013 Jun 1;12(6):414-21. doi: 10.1016/j.dnarep.2013.03.004. Epub 2013 Apr 11. DNA Repair (Amst). 2013. PMID: 23583337 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
