SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair
- PMID: 20157594
- PMCID: PMC2815768
- DOI: 10.18632/aging.100011
SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair
Abstract
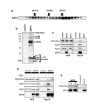
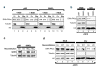
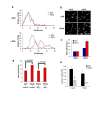
The Sir2 chromatin regulatory factor links maintenance of genomic stability to life span extension in yeast. The mammalian Sir2 family member SIRT6 has been proposed to have analogous functions, because SIRT6-deficiency leads to shortened life span and an aging-like degenerative phenotype in mice, and SIRT6 knockout cells exhibit genomic instability and DNA damage hypersensitivity. However, the molecular mechanisms underlying these defects are not fully understood. Here, we show that SIRT6 forms a macromolecular complex with the DNA double-strand break (DSB) repair factor DNA-PK (DNA-dependent protein kinase) and promotes DNA DSB repair. In response to DSBs, SIRT6 associates dynamically with chromatin and is necessary for an acute decrease in global cellular acetylation levels on histone H3 Lysine 9. Moreover, SIRT6 is required for mobilization of the DNA-PK catalytic subunit (DNA-PKcs) to chromatin in response to DNA damage and stabilizes DNA-PKcs at chromatin adjacent to an induced site-specific DSB. Abrogation of these SIRT6 activities leads to impaired resolution of DSBs. Together, these findings elucidate a mechanism whereby regulation of dynamic interaction of a DNA repair factor with chromatin impacts on the efficiency of repair, and establish a link between chromatin regulation, DNA repair, and a mammalian Sir2 factor.
Keywords: DNA damage; DNA repair; SIRT6; Sir2; aging; genomic stability.
Conflict of interest statement
The authors of this manuscript have no conflict of interests to declare.
Figures
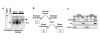


Comment in
-
Sirtuins at the breaking point: SIRT6 in DNA repair.Aging (Albany NY). 2009 Jan 20;1(1):12-6. doi: 10.18632/aging.100014. Aging (Albany NY). 2009. PMID: 20157593 Free PMC article. No abstract available.
Similar articles
-
Histone H2A.Z controls a critical chromatin remodeling step required for DNA double-strand break repair.Mol Cell. 2012 Dec 14;48(5):723-33. doi: 10.1016/j.molcel.2012.09.026. Epub 2012 Oct 30. Mol Cell. 2012. PMID: 23122415 Free PMC article.
-
Sirtuins at the breaking point: SIRT6 in DNA repair.Aging (Albany NY). 2009 Jan 20;1(1):12-6. doi: 10.18632/aging.100014. Aging (Albany NY). 2009. PMID: 20157593 Free PMC article. No abstract available.
-
DNA-PK is activated by nucleosomes and phosphorylates H2AX within the nucleosomes in an acetylation-dependent manner.Nucleic Acids Res. 2003 Dec 1;31(23):6819-27. doi: 10.1093/nar/gkg921. Nucleic Acids Res. 2003. PMID: 14627815 Free PMC article.
-
Chromatin regulation and genome maintenance by mammalian SIRT6.Trends Biochem Sci. 2011 Jan;36(1):39-46. doi: 10.1016/j.tibs.2010.07.009. Epub 2010 Aug 21. Trends Biochem Sci. 2011. PMID: 20729089 Free PMC article. Review.
-
The Chromatin Landscape around DNA Double-Strand Breaks in Yeast and Its Influence on DNA Repair Pathway Choice.Int J Mol Sci. 2023 Feb 7;24(4):3248. doi: 10.3390/ijms24043248. Int J Mol Sci. 2023. PMID: 36834658 Free PMC article. Review.
Cited by
-
SIRT6 mono-ADP ribosylates KDM2A to locally increase H3K36me2 at DNA damage sites to inhibit transcription and promote repair.Aging (Albany NY). 2020 Jun 25;12(12):11165-11184. doi: 10.18632/aging.103567. Epub 2020 Jun 25. Aging (Albany NY). 2020. PMID: 32584788 Free PMC article.
-
Targeting the Interplay between HDACs and DNA Damage Repair for Myeloma Therapy.Int J Mol Sci. 2021 Sep 27;22(19):10406. doi: 10.3390/ijms221910406. Int J Mol Sci. 2021. PMID: 34638744 Free PMC article. Review.
-
Sirtuins in hematological aging and malignancy.Crit Rev Oncog. 2013;18(6):531-47. doi: 10.1615/critrevoncog.2013010187. Crit Rev Oncog. 2013. PMID: 24579733 Free PMC article. Review.
-
JNK Phosphorylates SIRT6 to Stimulate DNA Double-Strand Break Repair in Response to Oxidative Stress by Recruiting PARP1 to DNA Breaks.Cell Rep. 2016 Sep 6;16(10):2641-2650. doi: 10.1016/j.celrep.2016.08.006. Epub 2016 Aug 25. Cell Rep. 2016. PMID: 27568560 Free PMC article.
-
SIRT6 in Aging, Metabolism, Inflammation and Cardiovascular Diseases.Aging Dis. 2022 Dec 1;13(6):1787-1822. doi: 10.14336/AD.2022.0413. eCollection 2022 Dec 1. Aging Dis. 2022. PMID: 36465178 Free PMC article. Review.
References
-
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles - a cause of aging in yeast. Cell. 1997;91:1033–1042. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
