Development and validation of a transcreener assay for detection of AMP- and GMP-producing enzymes
- PMID: 20158441
- PMCID: PMC2894640
- DOI: 10.1089/adt.2009.0254
Development and validation of a transcreener assay for detection of AMP- and GMP-producing enzymes
Abstract
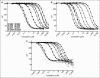
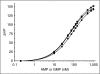
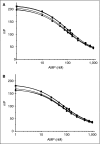
Screening of AMP- and GMP-producing enzymes such as phosphodiesterases (PDEs), ligases, and synthetases would be simplified by the ability to directly detect unmodified nucleoside monophosphates. To address this need, we developed polyclonal and monoclonal antibodies that recognize AMP and GMP with nanomolar sensitivity and high selectivity vs. the corresponding triphosphate and 3',5'-cyclic monophosphate nucleotides that serve as substrates for many enzymes in these classes. One of these antibodies was used to develop a Transcreener AMP/GMP assay with a far red fluorescence polarization (FP) readout. This polyclonal antibody exhibited extremely high selectivity, with IC(50) ratios of 6,000 for ATP/AMP, 3,810 for cAMP/AMP, and 6,970 for cGMP/GMP. Standard curves mimicking enzymatic conversion of cAMP, cGMP, and ATP to the corresponding monophosphates yielded Z' values of >0.85 at 10% conversion. The assay reagents were shown to be stable for 24 h at room temperature, both before and after dispensing. The Transcreener AMP/GMP FP assay was used for enzymatic detection of cGMP- and cAMP-dependent PDEs 4A1A, 3A, and 9A2 and ATP-dependent ligases, acetyl CoA synthetase, and ubiquitin- activating enzyme (UBE1). Shifts of >100 mP were observed in the linear part of the progress curves for all enzymes tested, and the PDE isoforms exhibited the expected substrate and inhibitor selectivity. These studies demonstrate that direct immunodetection of AMP and GMP is a flexible, robust enzyme assay method for diverse AMP- and GMP-producing enzymes. Moreover, it eliminates many of the shortcomings of other methods including the need for fluorescently labeled substrates, the low signal:background inherent in substrate depletion assays, and the potential for interference with coupling enzymes.
Figures

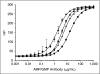




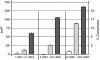
 ); the signal due to corresponding UBE1 quantities without UbcH2 (
); the signal due to corresponding UBE1 quantities without UbcH2 ( ); and the signal with this same concentration UbcH2 absent UBE1 (□).
); and the signal with this same concentration UbcH2 absent UBE1 (□).Similar articles
-
Rapid regulation of PDE-2 and PDE-4 cyclic AMP phosphodiesterase activity following ligation of the T cell antigen receptor on thymocytes: analysis using the selective inhibitors erythro-9-(2-hydroxy-3-nonyl)-adenine (EHNA) and rolipram.Cell Signal. 1996 Feb;8(2):97-110. doi: 10.1016/0898-6568(95)02032-2. Cell Signal. 1996. PMID: 8730511
-
Cyclic nucleotide phosphodiesterases (PDEs) in human osteoblastic cells; the effect of PDE inhibition on cAMP accumulation.Cell Mol Biol Lett. 2005;10(2):305-19. Cell Mol Biol Lett. 2005. PMID: 16010295
-
Role of cyclic AMP- and cyclic GMP-phosphodiesterases in the control of cyclic nucleotide levels and smooth muscle tone in rat isolated aorta. A study with selective inhibitors.Biochem Pharmacol. 1987 Nov 15;36(22):3965-72. doi: 10.1016/0006-2952(87)90465-5. Biochem Pharmacol. 1987. PMID: 2825708
-
Cyclic GMP and regulation of cyclic nucleotide hydrolysis.Adv Pharmacol. 1994;26:87-114. doi: 10.1016/s1054-3589(08)60052-6. Adv Pharmacol. 1994. PMID: 8038108 Review.
-
[Phosphodiesterases of cyclic GMP].Postepy Hig Med Dosw. 2001;55(5):611-27. Postepy Hig Med Dosw. 2001. PMID: 11795198 Review. Polish.
Cited by
-
A pathophysiological role of PDE3 in allergic airway inflammation.JCI Insight. 2018 Jan 25;3(2):e94888. doi: 10.1172/jci.insight.94888. eCollection 2018 Jan 25. JCI Insight. 2018. PMID: 29367458 Free PMC article.
-
Fluorescence polarization assays in small molecule screening.Expert Opin Drug Discov. 2011 Jan;6(1):17-32. doi: 10.1517/17460441.2011.537322. Expert Opin Drug Discov. 2011. PMID: 22328899 Free PMC article.
-
Cyclic Guanosine Monophosphate-Adenosine Monophosphate Synthase (cGAS), a Multifaceted Platform of Intracellular DNA Sensing.Front Immunol. 2021 Feb 23;12:637399. doi: 10.3389/fimmu.2021.637399. eCollection 2021. Front Immunol. 2021. PMID: 33708225 Free PMC article. Review.
-
Homogeneous single-label cGMP detection platform for the functional study of nitric oxide-sensitive (soluble) guanylyl cyclases and cGMP-specific phosphodiesterases.Sci Rep. 2020 Oct 15;10(1):17469. doi: 10.1038/s41598-020-74611-x. Sci Rep. 2020. PMID: 33060787 Free PMC article.
-
Development of a Rapid Mass Spectrometric Determination of AMP and Cyclic AMP for PDE3 Activity Study: Application and Computational Analysis for Evaluating the Effect of a Novel 2-oxo-1,2-dihydropyridine-3-carbonitrile Derivative as PDE-3 Inhibitor.Molecules. 2020 Apr 15;25(8):1817. doi: 10.3390/molecules25081817. Molecules. 2020. PMID: 32326556 Free PMC article.
References
-
- Lowery RG. Kleman-Leyer K. Transcreener: screening enzymes involved in covalent regulation. Expert Opin Ther Targets. 2006;10:179–190. - PubMed
-
- Hong L. Quinn CM. Jia Y. Evaluating the utility of the HTRF Transcreener ADP assay technology: a comparison with the standard HTRF assay technology. Anal Biochem. 2009;391:31–38. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

