The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease
- PMID: 20158612
- PMCID: PMC3934651
- DOI: 10.1111/j.1365-2958.2010.07070.x
The IbpA and IbpB small heat-shock proteins are substrates of the AAA+ Lon protease
Abstract
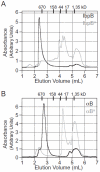
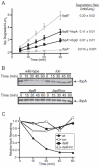
Small heat-shock proteins (sHSPs) are a widely conserved family of molecular chaperones, all containing a conserved alpha-crystallin domain flanked by variable N- and C-terminal tails. We report that IbpA and IbpB, the sHSPs of Escherichia coli, are substrates for the AAA+ Lon protease. This ATP-fueled enzyme degraded purified IbpA substantially more slowly than purified IbpB, and we demonstrate that this disparity is a consequence of differences in maximal Lon degradation rates and not in substrate affinity. Interestingly, however, IbpB stimulated Lon degradation of IbpA both in vitro and in vivo. Furthermore, although the variable N- and C-terminal tails of the Ibps were dispensable for proteolytic recognition, these tails contain critical determinants that control the maximal rate of Lon degradation. Finally, we show that E. coli Lon degrades variants of human alpha-crystallin, indicating that Lon recognizes conserved determinants in the folded alpha-crystallin domain itself. These results suggest a novel mode for Lon substrate recognition and provide a highly suggestive link between the degradation and sHSP branches of the protein quality-control network.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

