Computational approaches for detecting protein complexes from protein interaction networks: a survey
- PMID: 20158874
- PMCID: PMC2822531
- DOI: 10.1186/1471-2164-11-S1-S3
Computational approaches for detecting protein complexes from protein interaction networks: a survey
Abstract
Background: Most proteins form macromolecular complexes to perform their biological functions. However, experimentally determined protein complex data, especially of those involving more than two protein partners, are relatively limited in the current state-of-the-art high-throughput experimental techniques. Nevertheless, many techniques (such as yeast-two-hybrid) have enabled systematic screening of pairwise protein-protein interactions en masse. Thus computational approaches for detecting protein complexes from protein interaction data are useful complements to the limited experimental methods. They can be used together with the experimental methods for mapping the interactions of proteins to understand how different proteins are organized into higher-level substructures to perform various cellular functions.
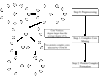
Results: Given the abundance of pairwise protein interaction data from high-throughput genome-wide experimental screenings, a protein interaction network can be constructed from protein interaction data by considering individual proteins as the nodes, and the existence of a physical interaction between a pair of proteins as a link. This binary protein interaction graph can then be used for detecting protein complexes using graph clustering techniques. In this paper, we review and evaluate the state-of-the-art techniques for computational detection of protein complexes, and discuss some promising research directions in this field.
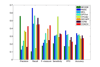
Conclusions: Experimental results with yeast protein interaction data show that the interaction subgraphs discovered by various computational methods matched well with actual protein complexes. In addition, the computational approaches have also improved in performance over the years. Further improvements could be achieved if the quality of the underlying protein interaction data can be considered adequately to minimize the undesirable effects from the irrelevant and noisy sources, and the various biological evidences can be better incorporated into the detection process to maximize the exploitation of the increasing wealth of biological knowledge available.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

