Differences in the transactivation domains of p53 family members: a computational study
- PMID: 20158876
- PMCID: PMC2822533
- DOI: 10.1186/1471-2164-11-S1-S5
Differences in the transactivation domains of p53 family members: a computational study
Abstract
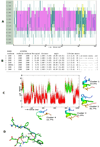
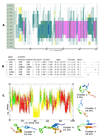
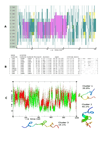
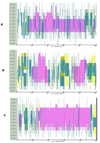
The N terminal transactivation domain of p53 is regulated by ligases and coactivator proteins. The functional conformation of this region appears to be an alpha helix which is necessary for its appropriate interactions with several proteins including MDM2 and p300. Folding simulation studies have been carried out to examine the propensity and stability of this region and are used to understand the differences between the family members with the ease of helix formation following the order p53 > p73 > p63. It is clear that hydrophobic clusters control the kinetics of helix formation, while electrostatic interactions control the thermodynamic stability of the helix. Differences in these interactions between the family members may partially account for the differential binding to, and regulation by, MDM2 (and MDMX). Phosphorylations of the peptides further modulate the stability of the helix and control associations with partner proteins.
Figures
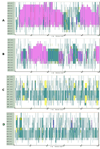
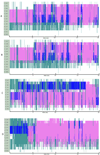
Similar articles
-
Interaction of regulators Mdm2 and Mdmx with transcription factors p53, p63 and p73.Cell Cycle. 2010 Nov 15;9(22):4584-91. doi: 10.4161/cc.9.22.13871. Epub 2010 Nov 15. Cell Cycle. 2010. PMID: 21088494
-
Molecular basis of the interactions between the p73 N terminus and p300: effects on transactivation and modulation by phosphorylation.Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3142-7. doi: 10.1073/pnas.0900383106. Epub 2009 Feb 13. Proc Natl Acad Sci U S A. 2009. PMID: 19218448 Free PMC article.
-
Conformational stability and activity of p73 require a second helix in the tetramerization domain.Cell Death Differ. 2009 Dec;16(12):1582-9. doi: 10.1038/cdd.2009.139. Epub 2009 Sep 18. Cell Death Differ. 2009. PMID: 19763140
-
The functional domains in p53 family proteins exhibit both common and distinct properties.Cell Death Differ. 2006 Jun;13(6):890-7. doi: 10.1038/sj.cdd.4401904. Cell Death Differ. 2006. PMID: 16543939 Review. No abstract available.
-
Structural diversity of p63 and p73 isoforms.Cell Death Differ. 2022 May;29(5):921-937. doi: 10.1038/s41418-022-00975-4. Epub 2022 Mar 21. Cell Death Differ. 2022. PMID: 35314772 Free PMC article. Review.
Cited by
-
Cancer-Associated Mutations Perturb the Disordered Ensemble and Interactions of the Intrinsically Disordered p53 Transactivation Domain.J Mol Biol. 2021 Jul 23;433(15):167048. doi: 10.1016/j.jmb.2021.167048. Epub 2021 May 11. J Mol Biol. 2021. PMID: 33984364 Free PMC article.
-
Tumor-Associated Antigen xCT and Mutant-p53 as Molecular Targets for New Combinatorial Antitumor Strategies.Cells. 2021 Jan 8;10(1):108. doi: 10.3390/cells10010108. Cells. 2021. PMID: 33430127 Free PMC article. Review.
-
Thirty years of molecular dynamics simulations on posttranslational modifications of proteins.Phys Chem Chem Phys. 2022 Nov 9;24(43):26371-26397. doi: 10.1039/d2cp02883b. Phys Chem Chem Phys. 2022. PMID: 36285789 Free PMC article. Review.
-
Molecular dynamic simulation insights into the normal state and restoration of p53 function.Int J Mol Sci. 2012;13(8):9709-9740. doi: 10.3390/ijms13089709. Epub 2012 Aug 3. Int J Mol Sci. 2012. PMID: 22949826 Free PMC article. Review.
-
Structural Evolution and Dynamics of the p53 Proteins.Cold Spring Harb Perspect Med. 2017 Apr 3;7(4):a028308. doi: 10.1101/cshperspect.a028308. Cold Spring Harb Perspect Med. 2017. PMID: 27091942 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

