Genomic features defining exonic variants that modulate splicing
- PMID: 20158892
- PMCID: PMC2872880
- DOI: 10.1186/gb-2010-11-2-r20
Genomic features defining exonic variants that modulate splicing
Abstract
Background: Single point mutations at both synonymous and non-synonymous positions within exons can have severe effects on gene function through disruption of splicing. Predicting these mutations in silico purely from the genomic sequence is difficult due to an incomplete understanding of the multiple factors that may be responsible. In addition, little is known about which computational prediction approaches, such as those involving exonic splicing enhancers and exonic splicing silencers, are most informative.
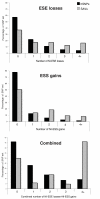
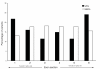
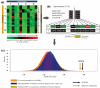
Results: We assessed the features of single-nucleotide genomic variants verified to cause exon skipping and compared them to a large set of coding SNPs common in the human population, which are likely to have no effect on splicing. Our findings implicate a number of features important for their ability to discriminate splice-affecting variants, including the naturally occurring density of exonic splicing enhancers and exonic splicing silencers of the exon and intronic environment, extensive changes in the number of predicted exonic splicing enhancers and exonic splicing silencers, proximity to the splice junctions and evolutionary constraint of the region surrounding the variant. By extending this approach to additional datasets, we also identified relevant features of variants that cause increased exon inclusion and ectopic splice site activation.
Conclusions: We identified a number of features that have statistically significant representation among exonic variants that modulate splicing. These analyses highlight putative mechanisms responsible for splicing outcome and emphasize the role of features important for exon definition. We developed a web-tool, Skippy, to score coding variants for these relevant splice-modulating features.
Figures

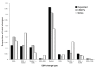

Similar articles
-
Vulnerable exons, like ACADM exon 5, are highly dependent on maintaining a correct balance between splicing enhancers and silencers.Hum Mutat. 2022 Feb;43(2):253-265. doi: 10.1002/humu.24321. Epub 2021 Dec 30. Hum Mutat. 2022. PMID: 34923709
-
Computational analysis of splicing errors and mutations in human transcripts.BMC Genomics. 2008 Jan 14;9:13. doi: 10.1186/1471-2164-9-13. BMC Genomics. 2008. PMID: 18194514 Free PMC article.
-
MutPred Splice: machine learning-based prediction of exonic variants that disrupt splicing.Genome Biol. 2014 Jan 13;15(1):R19. doi: 10.1186/gb-2014-15-1-r19. Genome Biol. 2014. PMID: 24451234 Free PMC article.
-
Rules and tools to predict the splicing effects of exonic and intronic mutations.Wiley Interdiscip Rev RNA. 2018 Jan;9(1). doi: 10.1002/wrna.1451. Epub 2017 Sep 26. Wiley Interdiscip Rev RNA. 2018. PMID: 28949076 Review.
-
Searching for splicing motifs.Adv Exp Med Biol. 2007;623:85-106. doi: 10.1007/978-0-387-77374-2_6. Adv Exp Med Biol. 2007. PMID: 18380342 Review.
Cited by
-
Computational identification of deleterious synonymous variants in human genomes using a feature-based approach.BMC Med Genomics. 2019 Jan 31;12(Suppl 1):12. doi: 10.1186/s12920-018-0455-6. BMC Med Genomics. 2019. PMID: 30704475 Free PMC article.
-
Lineage-specific splicing of a brain-enriched alternative exon promotes glioblastoma progression.J Clin Invest. 2014 Jul;124(7):2861-76. doi: 10.1172/JCI68836. Epub 2014 May 27. J Clin Invest. 2014. PMID: 24865424 Free PMC article.
-
Molecular genetic analysis of the PLP1 gene in 38 families with PLP1-related disorders: identification and functional characterization of 11 novel PLP1 mutations.Orphanet J Rare Dis. 2011 Jun 16;6:40. doi: 10.1186/1750-1172-6-40. Orphanet J Rare Dis. 2011. PMID: 21679407 Free PMC article.
-
Homozygosity mapping reveals novel and known mutations in Pakistani families with inherited retinal dystrophies.Sci Rep. 2015 May 6;5:9965. doi: 10.1038/srep09965. Sci Rep. 2015. PMID: 25943428 Free PMC article.
-
Exonic CLDN16 mutations associated with familial hypomagnesemia with hypercalciuria and nephrocalcinosis can induce deleterious mRNA alterations.BMC Med Genet. 2019 Jan 8;20(1):6. doi: 10.1186/s12881-018-0713-7. BMC Med Genet. 2019. PMID: 30621608 Free PMC article.
References
-
- Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

