The molecular determinants of the increased reduction potential of the rubredoxin domain of rubrerythrin relative to rubredoxin
- PMID: 20159152
- PMCID: PMC2820635
- DOI: 10.1016/j.bpj.2009.11.006
The molecular determinants of the increased reduction potential of the rubredoxin domain of rubrerythrin relative to rubredoxin
Abstract
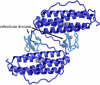
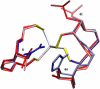
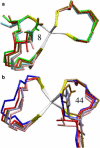
Based on the crystal structures, three possible sequence determinants have been suggested as the cause of a 285 mV increase in reduction potential of the rubredoxin domain of rubrerythrin over rubredoxin by modulating the polar environment around the redox site. Here, electrostatic calculations of crystal structures of rubredoxin and rubrerythrin and molecular dynamics simulations of rubredoxin wild-type and mutants are used to elucidate the contributions to the increased reduction potential. Asn(160) and His(179) in rubrerythrin versus valines in rubredoxins are predicted to be the major contributors, as the polar side chains contribute significantly to the electrostatic potential in the redox site region. The mutant simulations show both side chains rotating on a nanosecond timescale between two conformations with different electrostatic contributions. Reduction also causes a change in the reduction energy that is consistent with a linear response due to the interesting mechanism of shifting the relative populations of the two conformations. In addition to this, a simulation of a triple mutant indicates the side-chain rotations are approximately anticorrelated so whereas one is in the high potential conformation, the other is in the low potential conformation. However, Ala(176) in rubrerythrin versus a leucine in rubredoxin is not predicted to be a large contributor, because the solvent accessibility increases only slightly in mutant simulations and because it is buried in the interface of the rubrerythrin homodimer.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Johnson D.C., Dean D.R., Johnson M.K. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. - PubMed
-
- Meyer J. Iron-sulfur protein folds, iron-sulfur chemistry, and evolution. J. Biol. Inorg. Chem. 2008;13:157–170. - PubMed
-
- Beinert H., Meyer J., Lill R. Elsevier; Amsterdam, The Netherlands: 2004. Iron-sulfur proteins. In Encyclopedia of Biological Chemistry, Vol. 2. 482–489.
-
- Cammack R. Iron-sulfur cluster in enzymes: themes and variations. Adv. Inorg. Chem. 1992;38:281–322.
-
- Beinert H., Holm R.H., Münck E. Iron-sulfur clusters: nature's modular, multipurpose structures. Science. 1997;277:653–659. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

