Free energy profile of RNA hairpins: a molecular dynamics simulation study
- PMID: 20159159
- PMCID: PMC2820654
- DOI: 10.1016/j.bpj.2009.10.040
Free energy profile of RNA hairpins: a molecular dynamics simulation study
Abstract
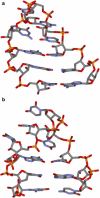
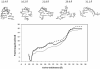
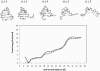
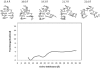
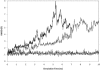
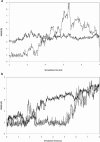
RNA hairpin loops are one of the most abundant secondary structure elements and participate in RNA folding and protein-RNA recognition. To characterize the free energy surface of RNA hairpin folding at an atomic level, we calculated the potential of mean force (PMF) as a function of the end-to-end distance, by using umbrella sampling simulations in explicit solvent. Two RNA hairpins containing tetraloop cUUCGg and cUUUUg are studied with AMBER ff99 and CHARMM27 force fields. Experimentally, the UUCG hairpin is known to be significantly more stable than UUUU. In this study, the calculations using AMBER force field give a qualitatively correct description for the folding of two RNA hairpins, as the calculated PMF confirms the global stability of the folded structures and the resulting relative folding free energy is in quantitative agreement with the experimental result. The hairpin stabilities are also correctly differentiated by the more rapid molecular mechanics-Poisson Boltzmann-surface area approach, but the relative free energy estimated from this method is overestimated. The free energy profile shows that the native state basin and the unfolded state plateau are separated by a wide shoulder region, which samples a variety of native-like structures with frayed terminal basepair. The calculated PMF lacks major barriers that are expected near the transition regions, and this is attributed to the limitation of the 1-D reaction coordinate. The PMF results are compared with other studies of small RNA hairpins using kinetics method and coarse grained models. The two RNA hairpins described by CHARMM27 are significantly more deformable than those represented by AMBER. Compared with the AMBER results, the CHARMM27 calculated DeltaG(fold) for the UUUU tetraloop is in better agreement with the experimental results. However, the CHARMM27 calculation does not confirm the global stability of the experimental UUCG structure; instead, the extended conformations are predicted to be thermodynamically stable in solution. This finding is further supported by separate unrestrained CHARMM27 simulations, in which the UUCG hairpin unfolds spontaneously within 10 ns. The instability of the UUCG hairpin originates from the loop region, and propagates to the stem. The results of this study provide a molecular picture of RNA hairpin unfolding and reveal problems in the force field descriptions for the conformational energy of certain RNA hairpin.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Tinoco I., Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. - PubMed
-
- Proctor D.J., Ma H., Bevilacqua P.C. Folding thermodynamics and kinetics of YNMG RNA hairpins: specific incorporation of 8-bromoguanosine leads to stabilization by enhancement of the folding rate. Biochemistry. 2004;43:14004–14014. - PubMed
-
- Uhlenbeck O.C. Tetraloops and RNA folding. Nature. 1990;346:613–614. - PubMed
-
- Varani G. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 1995;24:379–404. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

