TEM-1 backbone dynamics-insights from combined molecular dynamics and nuclear magnetic resonance
- PMID: 20159160
- PMCID: PMC2820648
- DOI: 10.1016/j.bpj.2009.08.061
TEM-1 backbone dynamics-insights from combined molecular dynamics and nuclear magnetic resonance
Abstract
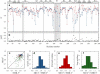
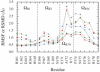
Dynamic properties of class A beta-lactamase TEM-1 are investigated from molecular dynamics (MD) simulations. Comparison of MD-derived order parameters with those obtained from model-free analysis of nuclear magnetic resonance (NMR) relaxation data shows high agreement for N-H moieties within alpha- and beta-secondary structures, but significant deviation for those in loops. This was expected, because motions slower than the protein global tumbling often take place in loop regions. As previously shown using NMR, TEM-1 is a highly ordered protein. Motions are observed within the Omega loop that could, upon substrate binding, stabilize E166 in a catalytically efficient position as the cavity between the protein core and the Omega loop is partially filled. The rigidity of active site residues is consistent with the enzyme high turnover number. MD data are also shown to be useful during the model selection step of model-free analysis: local N-H motions observed over the course of the trajectories help assess whether a peptide plan undergoes low or high amplitude motions on one or more timescales. This joint use of MD and NMR provides a better description of protein dynamics than would be possible using either technique alone.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures



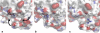
References
-
- Savard P.-Y., Gagné S.M. Backbone dynamics of TEM-1 determined by NMR: evidence for a highly ordered protein. Biochemistry. 2006;45:11414–11424. - PubMed
-
- Hermann J.C., Ridder L., Höltje H.D. Identification of Glu166 as the general base in the acylation reaction of class A β-lactamases through QM/MM modeling. J. Am. Chem. Soc. 2003;125:9590–9591. - PubMed
-
- Meroueh S.O., Fisher J.F., Mobashery S. Ab initio QM/MM study of class A β-lactamase acylation: dual participation of Glu166 and Lys73 in a concerted base promotion of Ser70. J. Am. Chem. Soc. 2005;127:15397–15407. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

