(Un)Folding mechanisms of the FBP28 WW domain in explicit solvent revealed by multiple rare event simulation methods
- PMID: 20159161
- PMCID: PMC2820652
- DOI: 10.1016/j.bpj.2009.10.039
(Un)Folding mechanisms of the FBP28 WW domain in explicit solvent revealed by multiple rare event simulation methods
Abstract
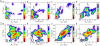
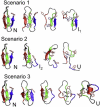
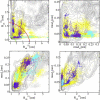
We report a numerical study of the (un)folding routes of the truncated FBP28 WW domain at ambient conditions using a combination of four advanced rare event molecular simulation techniques. We explore the free energy landscape of the native state, the unfolded state, and possible intermediates, with replica exchange molecular dynamics. Subsequent application of bias-exchange metadynamics yields three tentative unfolding pathways at room temperature. Using these paths to initiate a transition path sampling simulation reveals the existence of two major folding routes, differing in the formation order of the two main hairpins, and in hydrophobic side-chain interactions. Having established that the hairpin strand separation distances can act as reasonable reaction coordinates, we employ metadynamics to compute the unfolding barriers and find that the barrier with the lowest free energy corresponds with the most likely pathway found by transition path sampling. The unfolding barrier at 300 K is approximately 17 k(B)T approximately 42 kJ/mol, in agreement with the experimental unfolding rate constant. This work shows that combining several powerful simulation techniques provides a more complete understanding of the kinetic mechanism of protein folding.
Copyright 2010 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures
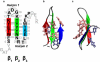
References
-
- Petrovich M., Jonsson A.L., Fersht A.R. Phi-analysis at the experimental limits: mechanism of β-hairpin formation. J. Mol. Biol. 2006;360:865–881. - PubMed
-
- Periole X., Allen L.R., Paci E. Probing the free energy landscape of the FBP28WW domain using multiple techniques. J. Comput. Chem. 2009;30:1059–1068. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

