Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded
- PMID: 20159598
- PMCID: PMC2858562
- DOI: 10.1016/j.devcel.2009.12.012
Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded
Abstract
The Hippo signaling pathway regulates organ size and tissue homeostasis from Drosophila to mammals. Central to this pathway is a kinase cascade wherein Hippo (Hpo), in complex with Salvador (Sav), phosphorylates and activates Warts (Wts), which in turn phosphorylates and inactivates the Yorkie (Yki) oncoprotein, known as the YAP coactivator in mammalian cells. The FERM domain proteins Merlin (Mer) and Expanded (Ex) are upstream components that regulate Hpo activity through unknown mechanisms. Here we identify Kibra as another upstream component of the Hippo signaling pathway. We show that Kibra functions together with Mer and Ex in a protein complex localized to the apical domain of epithelial cells, and that this protein complex regulates the Hippo kinase cascade via direct binding to Hpo and Sav. These results shed light on the mechanism of Ex and Mer function and implicate Kibra as a potential tumor suppressor with relevance to neurofibromatosis.
Copyright (c) 2010 Elsevier Inc. All rights reserved.
Figures

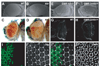
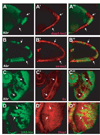
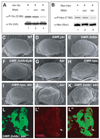



References
-
- Badouel C, Gardano L, Amin N, Garg A, Rosenfeld R, Le BT, McNeill H. The FERM-domain protein expanded regulates Hippo pathway activity via direct interactions with the transcriptional activator Yorkie. Dev. Cell. 2009;16:411–420. - PubMed
-
- Bennett FC, Harvey KF. Fat cadherin modulates organ size in Drosophila via the Salvador/Warts/Hippo signaling pathway. Curr. Biol. 2006;16:2101–2110. - PubMed
-
- Boedigheimer M, Laughon A. Expanded: a gene involved in the control of cell proliferation in imaginal discs. Development. 1993;118:1291–1301. - PubMed
-
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 2007;17:2054–2060. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

