A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis
- PMID: 20159959
- PMCID: PMC2816739
- DOI: 10.1101/gad.1878110
A highly coordinated cell wall degradation machine governs spore morphogenesis in Bacillus subtilis
Abstract
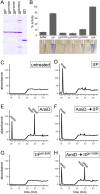
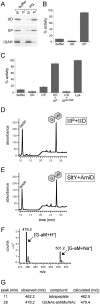
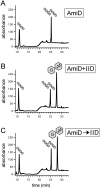
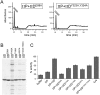
How proteins catalyze morphogenesis is an outstanding question in developmental biology. In bacteria, morphogenesis is intimately linked to remodeling the cell wall exoskeleton. Here, we investigate the mechanisms by which the mother cell engulfs the prospective spore during sporulation in Bacillus subtilis. A membrane-anchored protein complex containing two cell wall hydrolases plays a central role in this morphological process. We demonstrate that one of the proteins (SpoIIP) has both amidase and endopeptidase activities, such that it removes the stem peptides from the cell wall and cleaves the cross-links between them. We further show that the other protein (SpoIID) is the founding member of a new family of lytic transglycosylases that degrades the glycan strands of the peptidoglycan into disaccharide units. Importantly, we show that SpoIID binds the cell wall, but will only cleave the glycan strands after the stem peptides have been removed. Finally, we demonstrate that SpoIID also functions as an enhancer of SpoIIP activity. Thus, this membrane-anchored enzyme complex is endowed with complementary, sequential, and stimulatory activities. These activities provide a mechanism for processive cell wall degradation, supporting a model in which circumferentially distributed degradation machines function as motors pulling the mother cell membranes around the forespore.
Figures


Similar articles
-
SpoIID-mediated peptidoglycan degradation is required throughout engulfment during Bacillus subtilis sporulation.J Bacteriol. 2010 Jun;192(12):3174-86. doi: 10.1128/JB.00127-10. Epub 2010 Apr 9. J Bacteriol. 2010. PMID: 20382772 Free PMC article.
-
Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins.Mol Microbiol. 2007 Apr;64(1):139-52. doi: 10.1111/j.1365-2958.2007.05652.x. Mol Microbiol. 2007. PMID: 17376078
-
A SpoIID Homolog Cleaves Glycan Strands at the Chlamydial Division Septum.mBio. 2019 Jul 16;10(4):e01128-19. doi: 10.1128/mBio.01128-19. mBio. 2019. PMID: 31311880 Free PMC article.
-
Autolysins of Bacillus subtilis: multiple enzymes with multiple functions.Microbiology (Reading). 2000 Feb;146 ( Pt 2):249-262. doi: 10.1099/00221287-146-2-249. Microbiology (Reading). 2000. PMID: 10708363 Review. No abstract available.
-
Differentiation and the establishment of cell type during sporulation in Bacillus subtilis.Curr Opin Genet Dev. 1991 Oct;1(3):330-5. doi: 10.1016/s0959-437x(05)80296-5. Curr Opin Genet Dev. 1991. PMID: 1840889 Review.
Cited by
-
Substrate specificity of an elongation-specific peptidoglycan endopeptidase and its implications for cell wall architecture and growth of Vibrio cholerae.Mol Microbiol. 2013 Sep;89(5):949-62. doi: 10.1111/mmi.12323. Epub 2013 Jul 29. Mol Microbiol. 2013. PMID: 23834664 Free PMC article.
-
The SpoIIQ landmark protein has different requirements for septal localization and immobilization.Mol Microbiol. 2013 Sep;89(6):1053-68. doi: 10.1111/mmi.12333. Epub 2013 Aug 14. Mol Microbiol. 2013. PMID: 23859254 Free PMC article.
-
Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria.Mol Microbiol. 2016 Feb;99(4):700-18. doi: 10.1111/mmi.13258. Epub 2015 Nov 19. Mol Microbiol. 2016. PMID: 26507882 Free PMC article.
-
Milestones in Bacillus subtilis sporulation research.Microb Cell. 2020 Nov 27;8(1):1-16. doi: 10.15698/mic2021.01.739. Microb Cell. 2020. PMID: 33490228 Free PMC article. Review.
-
Shaping an Endospore: Architectural Transformations During Bacillus subtilis Sporulation.Annu Rev Microbiol. 2020 Sep 8;74:361-386. doi: 10.1146/annurev-micro-022520-074650. Epub 2020 Jul 13. Annu Rev Microbiol. 2020. PMID: 32660383 Free PMC article. Review.
References
-
- Blackburn NT, Clarke AJ. Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52:78–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
