Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ
- PMID: 20159972
- PMCID: PMC2857029
- DOI: 10.1074/jbc.M109.071688
Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ
Abstract
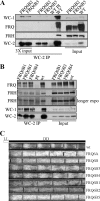
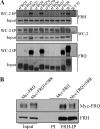
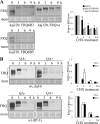
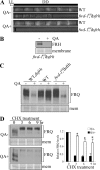
FREQUENCY (FRQ) is the central component of the Neurospora circadian clock. All FRQ proteins form the FFC complex with FRH (FRQ-interacting RNA helicase) that acts as the negative element in the circadian negative feedback loop by repressing frq mRNA levels. To understand the function of the FRQ-FRH interaction, we mapped and identified the minimal FRQ region that is required for FRQ-FRH interaction. We demonstrated that the FRQ-FRH complex formation is required for the interaction between FRQ and the White Collar Complex (WCC) and clock function. On the other hand, in the FRQ-FRH complex, FRQ is also required for the FRH-WCC interaction. Disruption of FRQ-FRH interaction or down-regulation of FRH results in hypophosphorylation, rapid degradation of FRQ, as well as low levels of WHITE COLLAR-1 and WHITE COLLAR-2. Furthermore, we showed that the rapid FRQ degradation in the absence of FRH is independent of FWD-1, the ubiquitin E3 ligase of FRQ under normal conditions, thus uncovering an alternative pathway for FRQ degradation.
Figures



Similar articles
-
FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock.Genetics. 2010 Feb;184(2):351-61. doi: 10.1534/genetics.109.111393. Epub 2009 Nov 30. Genetics. 2010. PMID: 19948888 Free PMC article.
-
Domains required for the interaction of the central negative element FRQ with its transcriptional activator WCC within the core circadian clock of Neurospora.J Biol Chem. 2023 Jul;299(7):104850. doi: 10.1016/j.jbc.2023.104850. Epub 2023 May 21. J Biol Chem. 2023. PMID: 37220856 Free PMC article.
-
Structure of the frequency-interacting RNA helicase: a protein interaction hub for the circadian clock.EMBO J. 2016 Aug 1;35(15):1707-19. doi: 10.15252/embj.201694327. Epub 2016 Jun 23. EMBO J. 2016. PMID: 27340124 Free PMC article.
-
Posttranslational regulation of Neurospora circadian clock by CK1a-dependent phosphorylation.Cold Spring Harb Symp Quant Biol. 2007;72:177-83. doi: 10.1101/sqb.2007.72.025. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419275 Review.
-
The molecular workings of the Neurospora biological clock.Novartis Found Symp. 2003;253:184-98; discussion 102-9, 198-202, 281-4. Novartis Found Symp. 2003. PMID: 14712922 Review.
Cited by
-
Circadian Interactomics: How Research Into Protein-Protein Interactions Beyond the Core Clock Has Influenced the Model of Circadian Timekeeping.J Biol Rhythms. 2021 Aug;36(4):315-328. doi: 10.1177/07487304211014622. Epub 2021 May 31. J Biol Rhythms. 2021. PMID: 34056936 Free PMC article. Review.
-
Data-driven modelling captures dynamics of the circadian clock of Neurospora crassa.PLoS Comput Biol. 2022 Aug 11;18(8):e1010331. doi: 10.1371/journal.pcbi.1010331. eCollection 2022 Aug. PLoS Comput Biol. 2022. PMID: 35951637 Free PMC article.
-
Natural Variation of the Circadian Clock in Neurospora.Adv Genet. 2017;99:1-37. doi: 10.1016/bs.adgen.2017.09.001. Epub 2017 Oct 12. Adv Genet. 2017. PMID: 29050553 Free PMC article. Review.
-
Disordered clock protein interactions and charge blocks turn an hourglass into a persistent circadian oscillator.Nat Commun. 2024 Apr 25;15(1):3523. doi: 10.1038/s41467-024-47761-z. Nat Commun. 2024. PMID: 38664421 Free PMC article.
-
Nonoptimal codon usage influences protein structure in intrinsically disordered regions.Mol Microbiol. 2015 Sep;97(5):974-87. doi: 10.1111/mmi.13079. Epub 2015 Jun 25. Mol Microbiol. 2015. PMID: 26032251 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

