Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme
- PMID: 20159977
- PMCID: PMC2852976
- DOI: 10.1074/jbc.M109.093583
Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II-degrading enzyme
Abstract
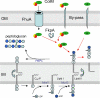
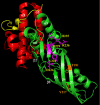
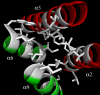
Colicin M inhibits Escherichia coli peptidoglycan synthesis through cleavage of its lipid-linked precursors. It has a compact structure, whereas other related toxins are organized in three independent domains, each devoted to a particular function: translocation through the outer membrane, receptor binding, and toxicity, from the N to the C termini, respectively. To establish whether colicin M displays such an organization despite its structural characteristics, protein dissection experiments were performed, which allowed us to delineate an independent toxicity domain encompassing exactly the C-terminal region conserved among colicin M-like proteins and covering about half of colicin M (residues 124-271). Surprisingly, the in vitro activity of the isolated domain was 45-fold higher than that of the full-length protein, suggesting a mechanism by which the toxicity of this domain is revealed following primary protein maturation. In vivo, the isolated toxicity domain appeared as toxic as the full-length protein under conditions where the reception and translocation steps were by-passed. Contrary to the full-length colicin M, the isolated domain did not require the presence of the periplasmic FkpA protein to be toxic under these conditions, demonstrating that FkpA is involved in the maturation process. Mutational analysis further identified five residues that are essential for cytotoxicity as well as in vitro lipid II-degrading activity: Asp-229, His-235, Asp-226, Tyr-228, and Arg-236. Most of these residues are surface-exposed and located relatively close to each other, hence suggesting they belong to the colicin M active site.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

