Chemotherapeutic drugs inhibit ribosome biogenesis at various levels
- PMID: 20159984
- PMCID: PMC2852979
- DOI: 10.1074/jbc.M109.074211
Chemotherapeutic drugs inhibit ribosome biogenesis at various levels
Abstract
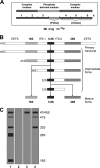
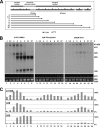
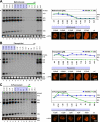
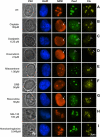
Drugs for cancer therapy belong to different categories of chemical substances. The cellular targets for the therapeutic efficacy are often not unambiguously identified. Here, we describe the process of ribosome biogenesis as a target of a large variety of chemotherapeutic drugs. We determined the inhibitory concentration of 36 chemotherapeutic drugs for transcription and processing of ribosomal RNA by in vivo labeling experiments. Inhibitory drug concentrations were correlated to the loss of nucleolar integrity. The synergism of drugs inhibiting ribosomal RNA synthesis at different levels was studied. Drugs inhibited ribosomal RNA synthesis either at the level of (i) rRNA transcription (e.g. oxaliplatin, doxorubicin, mitoxantrone, methotrexate), (ii) early rRNA processing (e.g. camptothecin, flavopiridol, roscovitine), or (iii) late rRNA processing (e.g. 5-fluorouracil, MG-132, homoharringtonine). Blockage of rRNA transcription or early rRNA processing steps caused nucleolar disintegration, whereas blockage of late rRNA processing steps left the nucleolus intact. Flavopiridol and 5-fluorouracil showed a strong synergism for inhibition of rRNA processing. We conclude that inhibition of ribosome biogenesis by chemotherapeutic drugs potentially may contribute to the efficacy of therapeutic regimens.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

