Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli
- PMID: 20160015
- PMCID: PMC2863545
- DOI: 10.1128/IAI.01174-09
Sequence analysis and characterization of a transferable hybrid plasmid encoding multidrug resistance and enabling zoonotic potential for extraintestinal Escherichia coli
Abstract
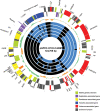
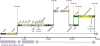
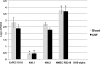
ColV plasmids of extraintestinal pathogenic Escherichia coli (ExPEC) encode a variety of fitness and virulence factors and have long been associated with septicemia and avian colibacillosis. These plasmids are found significantly more often in ExPEC, including ExPEC associated with human neonatal meningitis and avian colibacillosis, than in commensal E. coli. Here we describe pAPEC-O103-ColBM, a hybrid RepFIIA/FIB plasmid harboring components of the ColV pathogenicity island and a multidrug resistance (MDR)-encoding island. This plasmid is mobilizable and confers the ability to cause septicemia in chickens, the ability to cause bacteremia resulting in meningitis in the rat model of human disease, and the ability to resist the killing effects of multiple antimicrobial agents and human serum. The results of a sequence analysis of this and other ColV plasmids supported previous findings which indicated that these plasmid types arose from a RepFIIA/FIB plasmid backbone on multiple occasions. Comparisons of pAPEC-O103-ColBM with other sequenced ColV and ColBM plasmids indicated that there is a core repertoire of virulence genes that might contribute to the ability of some ExPEC strains to cause high-level bacteremia and meningitis in a rat model. Examination of a neonatal meningitis E. coli (NMEC) population revealed that approximately 58% of the isolates examined harbored ColV-type plasmids and that 26% of these plasmids had genetic contents similar to that of pAPEC-O103-ColBM. The linkage of the ability to confer MDR and the ability contribute to multiple forms of human and animal disease on a single plasmid presents further challenges for preventing and treating ExPEC infections.
Figures


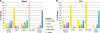
Similar articles
-
Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends.Foodborne Pathog Dis. 2013 Nov;10(11):916-32. doi: 10.1089/fpd.2013.1533. Epub 2013 Aug 20. Foodborne Pathog Dis. 2013. PMID: 23962019 Free PMC article. Review.
-
Complete DNA sequence of a ColBM plasmid from avian pathogenic Escherichia coli suggests that it evolved from closely related ColV virulence plasmids.J Bacteriol. 2006 Aug;188(16):5975-83. doi: 10.1128/JB.00204-06. J Bacteriol. 2006. PMID: 16885466 Free PMC article.
-
The plasmid of Escherichia coli strain S88 (O45:K1:H7) that causes neonatal meningitis is closely related to avian pathogenic E. coli plasmids and is associated with high-level bacteremia in a neonatal rat meningitis model.Infect Immun. 2009 Jun;77(6):2272-84. doi: 10.1128/IAI.01333-08. Epub 2009 Mar 23. Infect Immun. 2009. PMID: 19307211 Free PMC article.
-
Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease.Infect Immun. 2010 Aug;78(8):3412-9. doi: 10.1128/IAI.00347-10. Epub 2010 Jun 1. Infect Immun. 2010. PMID: 20515929 Free PMC article.
-
Escherichia coli from animal reservoirs as a potential source of human extraintestinal pathogenic E. coli.FEMS Immunol Med Microbiol. 2011 Jun;62(1):1-10. doi: 10.1111/j.1574-695X.2011.00797.x. Epub 2011 Mar 24. FEMS Immunol Med Microbiol. 2011. PMID: 21362060 Review.
Cited by
-
Human and avian extraintestinal pathogenic Escherichia coli: infections, zoonotic risks, and antibiotic resistance trends.Foodborne Pathog Dis. 2013 Nov;10(11):916-32. doi: 10.1089/fpd.2013.1533. Epub 2013 Aug 20. Foodborne Pathog Dis. 2013. PMID: 23962019 Free PMC article. Review.
-
Escherichia coli Isolated from Cases of Colibacillosis in Russian Poultry Farms (Perm Krai): Sensitivity to Antibiotics and Bacteriocins.Microorganisms. 2020 May 15;8(5):741. doi: 10.3390/microorganisms8050741. Microorganisms. 2020. PMID: 32429211 Free PMC article.
-
Genome evolution and the emergence of pathogenicity in avian Escherichia coli.Nat Commun. 2021 Feb 3;12(1):765. doi: 10.1038/s41467-021-20988-w. Nat Commun. 2021. PMID: 33536414 Free PMC article.
-
Zoonotic potential of multidrug-resistant extraintestinal pathogenic Escherichia coli obtained from healthy poultry carcasses in Salvador, Brazil.Braz J Infect Dis. 2013 Jan-Feb;17(1):54-61. doi: 10.1016/j.bjid.2012.09.004. Epub 2013 Jan 3. Braz J Infect Dis. 2013. PMID: 23290470 Free PMC article.
-
American Crows as Carriers of Extra Intestinal Pathogenic E. coli and Avian Pathogenic-Like E. coli and Their Potential Impact on a Constructed Wetland.Microorganisms. 2020 Oct 16;8(10):1595. doi: 10.3390/microorganisms8101595. Microorganisms. 2020. PMID: 33081240 Free PMC article.
References
-
- Aguero, M., G. de la Fuente, E. Vivaldi, and F. Cabello. 1989. ColV increases the virulence of Escherichia coli K1 strains in animal models of neonatal meningitis and urinary infection. Med. Microbiol. Immunol. 178:211-216. - PubMed
-
- Bonacorsi, S., and E. Bingen. 2005. Molecular epidemiology of Escherichia coli causing neonatal meningitis. Int. J. Med. Microbiol. 295:373-381. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

