Morphological analysis of 13 LMNA variants identified in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy
- PMID: 20160190
- PMCID: PMC2908895
- DOI: 10.1161/CIRCGENETICS.109.905422
Morphological analysis of 13 LMNA variants identified in a cohort of 324 unrelated patients with idiopathic or familial dilated cardiomyopathy
Abstract
Background: Mutations in the LMNA gene, encoding lamins A/C, represent a significant cause of dilated cardiomyopathy. We recently identified 18 protein-altering LMNA variants in a cohort of 324 unrelated patients with dilated cardiomyopathy. However, at least one family member with dilated cardiomyopathy in each of 6 pedigrees lacked the LMNA mutation (nonsegregation), whereas small sizes of 5 additional families precluded definitive determinations of segregation, raising questions regarding contributions by those variants to disease.
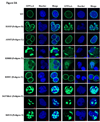
Methods and results: We have consequently expressed, in COS7 cells, GFP-prelamin A (GFPLaA) fusion constructs incorporating the 6 variants in pedigrees with nonsegregation (R101P, A318T, R388H, R399C, S437Hfsx1, and R654X), the 4 variants in pedigrees with unknown segregation (R89L, R166P [in 2 families], I210S, R471H), and 3 additional missense variants (R190Q, E203K, and L215P) that segregated with disease. Confocal immunofluorescence microscopy was used to characterize GFP-lamin A localization and nuclear morphology. Abnormal phenotypes were observed for 10 of 13 (77%) variants (R89L, R101P, R166P, R190Q, E203K, I210S, L215P, R388H, S437Hfsx1, and R654X), including 4 of 6 showing nonsegregation and 3 of 4 with uncertain segregation. All 7 variants affecting coil 1B and the lamin A-only mutation, R654X, exhibited membrane-bound GFP-lamin A aggregates and nuclear shape abnormalities. Unexpectedly, R388H largely restricted GFP-lamin A to the cytoplasm. Equally unexpected were unique streaked aggregates with S437Hfsx1 and giant aggregates with both S437Hfsx1 and R654X.
Conclusions: This work expands the recognized spectrum of lamin A localization abnormalities in dilated cardiomyopathy. It also provides evidence supporting pathogenicity of 10 of 13 tested LMNA variants, including some with uncertain or nonsegregation.
Conflict of interest statement
Figures



References
-
- Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nat Rev Genet. 2006;7:940–952. - PubMed
-
- Burke B, Stewart CL. The laminopathies: the functional architecture of the nucleus and its contribution to disease (*) Annu Rev Genomics Hum Genet. 2006;7:369–405. - PubMed
-
- Stuurman N, Sasse B, Fisher PA. Intermediate filament protein polymerization: molecular analysis of Drosophila nuclear lamin head-to-tail binding. J Struct Biol. 1996;117:1–15. - PubMed
-
- Zastrow MS, Vlcek S, Wilson KL. Proteins that bind A-type lamins: integrating isolated clues. J Cell Sci. 2004;117:979–987. - PubMed
-
- Holtz D, Tanaka RA, Hartwig J, McKeon F. The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell. 1989;59:969–977. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

