Different sets of QTLs influence fitness variation in yeast
- PMID: 20160707
- PMCID: PMC2835564
- DOI: 10.1038/msb.2010.1
Different sets of QTLs influence fitness variation in yeast
Abstract
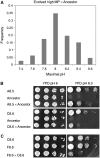
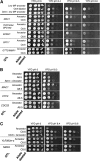
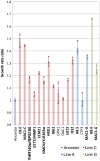
Most of the phenotypes in nature are complex and are determined by many quantitative trait loci (QTLs). In this study we identify gene sets that contribute to one important complex trait: the ability of yeast cells to survive under alkali stress. We carried out an in-lab evolution (ILE) experiment, in which we grew yeast populations under increasing alkali stress to enrich for beneficial mutations. The populations acquired different sets of affecting alleles, showing that evolution can provide alternative solutions to the same challenge. We measured the contribution of each allele to the phenotype. The sum of the effects of the QTLs was larger than the difference between the ancestor phenotype and the evolved strains, suggesting epistatic interactions between the QTLs. In parallel, a clinical isolated strain was used to map natural QTLs affecting growth at high pH. In all, 17 candidate regions were found. Using a predictive algorithm based on the distances in protein-interaction networks, candidate genes were defined and validated by gene disruption. Many of the QTLs found by both methods are not directly implied in pH homeostasis but have more general, and often regulatory, roles.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Concerted evolution of life stage performances signals recent selection on yeast nitrogen use.Mol Biol Evol. 2015 Jan;32(1):153-61. doi: 10.1093/molbev/msu285. Epub 2014 Oct 27. Mol Biol Evol. 2015. PMID: 25349282
-
Highly complete long-read genomes reveal pangenomic variation underlying yeast phenotypic diversity.Genome Res. 2023 May;33(5):729-740. doi: 10.1101/gr.277515.122. Epub 2023 May 1. Genome Res. 2023. PMID: 37127330 Free PMC article.
-
Causal inference of regulator-target pairs by gene mapping of expression phenotypes.BMC Genomics. 2006 May 24;7:125. doi: 10.1186/1471-2164-7-125. BMC Genomics. 2006. PMID: 16719927 Free PMC article.
-
High-throughput approaches to functional characterization of genetic variation in yeast.Curr Opin Genet Dev. 2022 Oct;76:101979. doi: 10.1016/j.gde.2022.101979. Epub 2022 Sep 5. Curr Opin Genet Dev. 2022. PMID: 36075138 Review.
-
QTL-based evidence for the role of epistasis in evolution.Genet Res. 2005 Oct;86(2):89-95. doi: 10.1017/S0016672305007780. Genet Res. 2005. PMID: 16356282 Review.
Cited by
-
Defining Molecular Basis for Longevity Traits in Natural Yeast Isolates.NPJ Aging Mech Dis. 2015;1:15001-. doi: 10.1038/npjamd.2015.1. Epub 2015 Sep 28. NPJ Aging Mech Dis. 2015. PMID: 27030810 Free PMC article.
-
Revealing the genetic structure of a trait by sequencing a population under selection.Genome Res. 2011 Jul;21(7):1131-8. doi: 10.1101/gr.116731.110. Epub 2011 Mar 21. Genome Res. 2011. PMID: 21422276 Free PMC article.
-
A natural variant of the essential host gene MMS21 restricts the parasitic 2-micron plasmid in Saccharomyces cerevisiae.Elife. 2020 Oct 16;9:e62337. doi: 10.7554/eLife.62337. Elife. 2020. PMID: 33063663 Free PMC article.
-
ATG18 and FAB1 are involved in dehydration stress tolerance in Saccharomyces cerevisiae.PLoS One. 2015 Mar 24;10(3):e0119606. doi: 10.1371/journal.pone.0119606. eCollection 2015. PLoS One. 2015. PMID: 25803831 Free PMC article.
-
From sequence to function: Insights from natural variation in budding yeasts.Biochim Biophys Acta. 2011 Oct;1810(10):959-66. doi: 10.1016/j.bbagen.2011.02.004. Epub 2011 Feb 12. Biochim Biophys Acta. 2011. PMID: 21320572 Free PMC article. Review.
References
-
- Abiola O, Angel JM, Avner P, Bachmanov AA, Belknap JK, Bennett B, Blankenhorn EP, Blizard DA, Bolivar V, Brockmann GA, Buck KJ, Bureau JF, Casley WL, Chesler EJ, Cheverud JM, Churchill GA, Cook M, Crabbe JC, Crusio WE, Darvasi A et al. (2003) The nature and identification of quantitative trait loci: a community's view. Nat Rev Genet 4: 911–916 - PMC - PubMed
-
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat Genet 33(Suppl): 228–237 - PubMed
-
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998) Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115–132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

