Coupled Flow-Structure-Biochemistry Simulations of Dynamic Systems of Blood Cells Using an Adaptive Surface Tracking Method
- PMID: 20160939
- PMCID: PMC2765665
- DOI: 10.1016/j.jfluidstructs.2009.02.002
Coupled Flow-Structure-Biochemistry Simulations of Dynamic Systems of Blood Cells Using an Adaptive Surface Tracking Method
Abstract
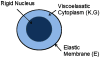
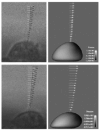
A method for the computation of low Reynolds number dynamic blood cell systems is presented. The specific system of interest here is interaction between cancer cells and white blood cells in an experimental flow system. Fluid dynamics, structural mechanics, six-degree-of freedom motion control and surface biochemistry analysis components are coupled in the context of adaptive octree-based grid generation. Analytical and numerical verification of the quasi-steady assumption for the fluid mechanics is presented. The capabilities of the technique are demonstrated by presenting several three-dimensional cell system simulations, including the collision/interaction between a cancer cell and an endothelium adherent polymorphonuclear leukocyte (PMN) cell in a shear flow.
Figures













References
-
- ACUSIM Software Inc. http://www.acusim.com/html/acusolve.html.
-
- Bell GI. Models for the specific adhesion adhesion of cells to cells. Science. 1978;200:618–627. - PubMed
-
- Belk DM, Maple RC. Automated assembly of structured grids for moving body problems. AIAA Paper 1995-1680.Proceedings of the 12th AIAA Computational Fluid Dynamics Conference; 1995.
-
- Brennen CE. Cavitation and Bubble Dynamics. Oxford University Press; New York, NY: 1995.
-
- Bongrand P, Bell GI. Cell-Cell Adhesion: Parameters and Possible Mechanisms. Marcel Dekker Inc.; New York, NY: 1984.
Grants and funding
LinkOut - more resources
Full Text Sources
