Characterization of protein secondary structure from NMR chemical shifts
- PMID: 20160946
- PMCID: PMC2766081
- DOI: 10.1016/j.pnmrs.2008.06.002
Characterization of protein secondary structure from NMR chemical shifts
Figures


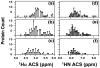
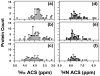
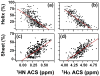
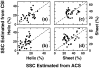
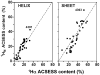

References
-
- Proctor WG, Yu FC. The dependence of a nuclear magnetic resonance frequency upon chemical compound. Phy Rev. 1950;77:717.
-
- Arnold JT, DSS, Packard ME. Chemical effects on nuclear-induction signals from organic compounds. J Chem Phys. 1951;19:507.
-
- Asakura T, Iwadate M, Demura M, Williamson MP. Structural analysis of silk with C-13 NMR chemical shift contour plots. Int J Biol Macromol. 1999;24:167–171. - PubMed
-
- Asakura T, Taoka K, Demura M, Williamson MP. The Relationship Between Amide Proton Chemical Shifts and Secondary Structure in Proteins. J Biomol NMR. 1995;6:227–236. - PubMed
-
- Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

