Multiple roles of mobile active center loops in the E1 component of the Escherichia coli pyruvate dehydrogenase complex - Linkage of protein dynamics to catalysis
- PMID: 20160956
- PMCID: PMC2759092
- DOI: 10.1016/j.molcatb.2009.04.008
Multiple roles of mobile active center loops in the E1 component of the Escherichia coli pyruvate dehydrogenase complex - Linkage of protein dynamics to catalysis
Abstract
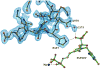
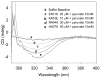
The region encompassing residues 401-413 on the E1 component of the pyruvate dehydrogenase multienzyme complex from Escherichia coli comprises a loop (the inner loop) which was not seen in the X-ray structure in the presence of thiamin diphosphate, the required cofactor for the enzyme. This loop is seen in the presence of a stable analogue of the pre-decarboxylation intermediate, the covalent adduct between the substrate analogue methyl acetylphosphonate and thiamin diphosphate, C2α-phosphonolactylthiamin diphosphate. It has been shown that the residue H407 and several other residues on this loop are required to reduce the mobility of the loop so electron density corresponding to it can be seen once the pre-decarboxylation intermediate is formed. Concomitantly, the loop encompassing residues 541-557 (the outer loop) appears to work in tandem with the inner loop and there is a hydrogen bond between the two loops ensuring their correlated motion. The inner loop was shown to: a) sequester the active center from carboligase side reactions; b) assist the interaction between the E1 and the E2 components, thereby affecting the overall reaction rate of the entire multienzyme complex; c) control substrate access to the active center. Using viscosity effects on kinetics it was shown that formation of the pre-decarboxylation intermediate is specifically affected by loop movement. A cysteine-less variant was created for the E1 component, onto which cysteines were substituted at selected loop positions. Introducing an electron spin resonance spin label and an (19)F NMR label onto these engineered cysteines, the loop mobility was examined: a) both methods suggested that in the absence of ligand, the loop exists in two conformations; b) line-shape analysis of the NMR signal at different temperatures, enabled estimation of the rate constant for loop movement, and this rate constant was found to be of the same order of magnitude as the turnover number for the enzyme under the same conditions. Furthermore, this analysis gave important insights into rate-limiting thermal loop dynamics. Overall, the results suggest that the dynamic properties correlate with catalytic events on the E1 component of the pyruvate dehydrogenase complex.
Figures






Similar articles
-
Efficient coupling of catalysis and dynamics in the E1 component of Escherichia coli pyruvate dehydrogenase multienzyme complex.Proc Natl Acad Sci U S A. 2008 Jan 29;105(4):1158-63. doi: 10.1073/pnas.0709328105. Epub 2008 Jan 23. Proc Natl Acad Sci U S A. 2008. PMID: 18216265 Free PMC article.
-
A dynamic loop at the active center of the Escherichia coli pyruvate dehydrogenase complex E1 component modulates substrate utilization and chemical communication with the E2 component.J Biol Chem. 2007 Sep 21;282(38):28106-16. doi: 10.1074/jbc.M704326200. Epub 2007 Jul 17. J Biol Chem. 2007. PMID: 17635929
-
A thiamin-bound, pre-decarboxylation reaction intermediate analogue in the pyruvate dehydrogenase E1 subunit induces large scale disorder-to-order transformations in the enzyme and reveals novel structural features in the covalently bound adduct.J Biol Chem. 2006 Jun 2;281(22):15296-303. doi: 10.1074/jbc.M600656200. Epub 2006 Mar 10. J Biol Chem. 2006. PMID: 16531404
-
Nuclear magnetic resonance approaches in the study of 2-oxo acid dehydrogenase multienzyme complexes--a literature review.Molecules. 2013 Sep 26;18(10):11873-903. doi: 10.3390/molecules181011873. Molecules. 2013. PMID: 24077172 Free PMC article. Review.
-
Characterization of enzyme motions by solution NMR relaxation dispersion.Acc Chem Res. 2008 Feb;41(2):214-21. doi: 10.1021/ar700132n. Epub 2008 Feb 19. Acc Chem Res. 2008. PMID: 18281945 Review.
Cited by
-
Structure and function of the catalytic domain of the dihydrolipoyl acetyltransferase component in Escherichia coli pyruvate dehydrogenase complex.J Biol Chem. 2014 May 30;289(22):15215-30. doi: 10.1074/jbc.M113.544080. Epub 2014 Apr 17. J Biol Chem. 2014. PMID: 24742683 Free PMC article.
-
Simulations of Pathogenic E1α Variants: Allostery and Impact on Pyruvate Dehydrogenase Complex-E1 Structure and Function.J Chem Inf Model. 2022 Jul 25;62(14):3463-3475. doi: 10.1021/acs.jcim.2c00630. Epub 2022 Jul 7. J Chem Inf Model. 2022. PMID: 35797142 Free PMC article.
-
The pyruvate dehydrogenase complexes: structure-based function and regulation.J Biol Chem. 2014 Jun 13;289(24):16615-23. doi: 10.1074/jbc.R114.563148. Epub 2014 May 5. J Biol Chem. 2014. PMID: 24798336 Free PMC article. Review.
-
Bifunctionality of the thiamin diphosphate cofactor: assignment of tautomeric/ionization states of the 4'-aminopyrimidine ring when various intermediates occupy the active sites during the catalysis of yeast pyruvate decarboxylase.J Am Chem Soc. 2012 Feb 29;134(8):3873-85. doi: 10.1021/ja211139c. Epub 2012 Feb 17. J Am Chem Soc. 2012. PMID: 22300533 Free PMC article.
-
Novel binding motif and new flexibility revealed by structural analyses of a pyruvate dehydrogenase-dihydrolipoyl acetyltransferase subcomplex from the Escherichia coli pyruvate dehydrogenase multienzyme complex.J Biol Chem. 2014 Oct 24;289(43):30161-76. doi: 10.1074/jbc.M114.592915. Epub 2014 Sep 10. J Biol Chem. 2014. PMID: 25210042 Free PMC article.
References
-
- Reed LJ. Acc, Chem, Res. 1974;7:40–47.
-
- Arjunan P, Nemeria N, Brunskill A, Chandrasekhar K, Sax M, Yan Y, Jordan F, Guest J, Furey W. Biochemistry. 2002;41:5213–5221. - PubMed
-
- Nemeria N, Arjunan P, Brunskill A, Sheibani F, Wei W, Yan Y, Zhang S, Jordan F, Furey W. Biochemistry. 2002;41:15459–15467. - PubMed
-
- Chiu C, Chung A, Barletta G, Jordan F. J Am Chem Soc. 1996;118:11026–11029.
-
- Pan K, Jordan F. Biochemistry. 1998;37:1357–1364. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
