Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins
- PMID: 20161395
- PMCID: PMC2782866
- DOI: 10.1016/j.pnmrs.2009.07.002
Recent Advances in the Application of Solution NMR Spectroscopy to Multi-Span Integral Membrane Proteins
Figures



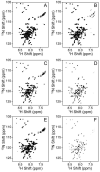
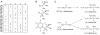
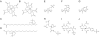



References
-
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Science. 1996;274:546. - PubMed
-
- Jimonet P, Jager R. Curr. Opin. Drug. Discov. Devel. 2004;7:325. - PubMed
-
- Schnur DM, Hermsmeier MA, Tebben AJ. J. Med. Chem. 2006;49:2000. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

