Temporal expression of chemokines dictates the hepatic inflammatory infiltrate in a murine model of schistosomiasis
- PMID: 20161726
- PMCID: PMC2817718
- DOI: 10.1371/journal.pntd.0000598
Temporal expression of chemokines dictates the hepatic inflammatory infiltrate in a murine model of schistosomiasis
Abstract
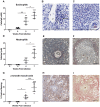
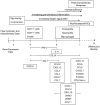
Schistosomiasis continues to be an important cause of parasitic morbidity and mortality world-wide. Determining the molecular mechanisms regulating the development of granulomas and fibrosis will be essential for understanding how schistosome antigens interact with the host environment. We report here the first whole genome microarray analysis of the murine liver during the progression of Schistosoma japonicum egg-induced granuloma formation and hepatic fibrosis. Our results reveal a distinct temporal relationship between the expression of chemokine subsets and the recruitment of cells to the infected liver. Genes up-regulated earlier in the response included T- and B-cell chemoattractants, reflecting the early recruitment of these cells illustrated by flow cytometry. The later phases of the response corresponded with peak recruitment of eosinophils, neutrophils, macrophages and myofibroblasts/hepatic stellate cells (HSCs) and the expression of chemokines with activity for these cells including CCL11 (eotaxin 1), members of the Monocyte-chemoattractant protein family (CCL7, CCL8, CCL12) and the Hepatic Stellate Cell/Fibrocyte chemoattractant CXCL1. Peak expression of macrophage chemoattractants (CCL6, CXCL14) and markers of alternatively activated macrophages (e.g. Retnla) during this later phase provides further evidence of a role for these cells in schistosome-induced pathology. Additionally, we demonstrate that CCL7 immunolocalises to the fibrotic zone of granulomas. Furthermore, striking up-regulation of neutrophil markers and the localisation of neutrophils and the neutrophil chemokine S100A8 to fibrotic areas suggest the involvement of neutrophils in S. japonicum-induced hepatic fibrosis. These results further our understanding of the immunopathogenic and, especially, chemokine signalling pathways that regulate the development of S. japonicum-induced granulomas and fibrosis and may provide correlative insight into the pathogenesis of other chronic inflammatory diseases of the liver where fibrosis is a common feature.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Burke ML, Jones MK, Gobert GN, Li YS, Ellis MK, et al. Immunopathogenesis of human schistosomiasis. Parasite Immunol. 2009;31:163–176. - PubMed
-
- Cheever AW. Comparison of pathologic changes in mammalian hosts infected with Schistosoma mansoni, S. japonicum and S. haematobium. Mem Inst Oswaldo Cruz. 1987;82(Suppl 4):39–45. - PubMed
-
- Hsu SY, Hsu HF, Davis JR, Lust GL. Comparative studies on the lesions caused by eggs of Schistosoma japonicum and Schistosoma mansoni in livers of albino mice and rhesus monkeys. Ann Trop Med Parasitol. 1972;66:89–97. - PubMed
-
- Warren KS, Grove DI, Pelley RP. The Schistosoma japonicum egg granuloma. II. Cellular composition, granuloma size, and immunologic concomitants. Am J Trop Med Hyg. 1978;27:271–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

