Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos
- PMID: 20161773
- PMCID: PMC2818844
- DOI: 10.1371/journal.pone.0009150
Histone H3 lysine 27 methylation asymmetry on developmentally-regulated promoters distinguish the first two lineages in mouse preimplantation embryos
Abstract
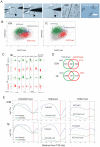
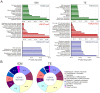
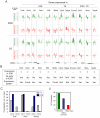
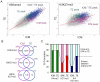
First lineage specification in the mammalian embryo leads to formation of the inner cell mass (ICM) and trophectoderm (TE), which respectively give rise to embryonic and extraembryonic tissues. We show here that this first differentiation event is accompanied by asymmetric distribution of trimethylated histone H3 lysine 27 (H3K27me3) on promoters of signaling and developmentally-regulated genes in the mouse ICM and TE. A genome-wide survey of promoter occupancy by H3K4me3 and H3K27me3 indicates that both compartments harbor promoters enriched in either modification, and promoters co-enriched in trimethylated H3K4 and H3K27 linked to developmental and signaling functions. The majority of H3K4/K27me3 co-enriched promoters are distinct between the two lineages, primarily due to differences in the distribution of H3K27me3. Derivation of embryonic stem cells leads to significant losses and gains of H3K4/K27me3 co-enriched promoters relative to the ICM, with distinct contributions of (de)methylation events on K4 and K27. Our results show histone trimethylation asymmetry on promoters in the first two developmental lineages, and highlight an epigenetic skewing associated with embryonic stem cell derivation.
Conflict of interest statement
Figures

References
-
- Rossant J, Tam PP. Blastocyst lineage formation, early embryonic asymmetries and axis patterning in the mouse. Development. 2009;136:701–713. - PubMed
-
- Albert M, Peters AH. Genetic and epigenetic control of early mouse development. Curr Opin Genet Dev. 2009;19:113–121. - PubMed
-
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14:R47–R58. - PubMed
-
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, et al. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

