Characteristics of Epstein-Barr virus envelope protein gp42
- PMID: 20162447
- PMCID: PMC2854865
- DOI: 10.1007/s11262-010-0455-x
Characteristics of Epstein-Barr virus envelope protein gp42
Abstract
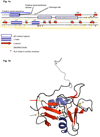
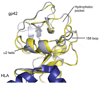
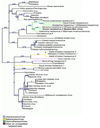
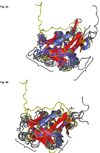
Epstein-Barr virus (EBV) glycoprotein 42 (gp42) is a membrane protein essential for fusion and entry of EBV into host B-lymphocytes. Gp42 is a member of the protein-fold family C-type lectin or lectin-like domains (CLECT or CTLD) and specifically is classified as a natural-killer receptor (NKR)-like CLECT. Literature review and phylogenetic comparison show that EBV gp42 shares a common structure with other NKR-like CLECTs and possibly with many viral CTLDs, but does not appear to exhibit some common binding characteristics of many CTLDs, such as features required for calcium binding. The flexible N-terminal region adjacent to the CTLD fold is important for binding to other EBV glycoproteins and for a cleavage site that is necessary for infection of host cells. From structural studies of gp42 unbound and bound to receptor and extensive mutational analysis, a general model of how gp42 triggers membrane fusion utilizing both the flexible N-terminal region and the CTLD domain has emerged.
Figures
References
-
- Rickinson A, Kieff E. In: Fields' Virology. Fields BN, Knipe DM, Howley PM, editors. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2656–2700.
-
- Henle W, Henle G. Ann Clin Lab Sci. 1974;4:109–114. - PubMed
-
- Kutok JL, Wang F. Annu Rev Pathol. 2006;1:375–404. - PubMed
-
- Maeda E, Akahane M, Kiryu S, Kato N, Yoshikawa T, Hayashi N, Aoki S, Minami M, Uozaki H, Fukayama M, Ohtomo K. Jpn J Radiol. 2009;27:4–19. - PubMed
-
- Rezk SA, Weiss LM. Hum Pathol. 2007;38:1293–1304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

