Discovery and characterization of a mammalian amyloid disaggregation activity
- PMID: 20162625
- PMCID: PMC2867023
- DOI: 10.1002/pro.363
Discovery and characterization of a mammalian amyloid disaggregation activity
Abstract
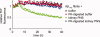
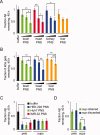
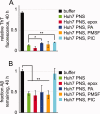
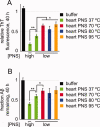
The formation of amyloid, a cross-beta-sheet fibrillar aggregate, is associated with a variety of aging-associated degenerative diseases. Herein, we report the existence of a mammalian amyloid disaggregase activity that is present in all tissues and cell types tested. Homogenates from mammalian tissues and cell lines are able to disaggregate amyloid fibrils composed of amyloid beta (A beta)(1-40) or the 8 kDa plasma gelsolin fragment. The mammalian disaggregase activity is sensitive to proteinase K digestion and can be uncoupled from proteolysis activity using a protease inhibitor cocktail. Amyloid disaggregation and proteolysis activities are remarkably resistant to changes in temperature and pH. Identification and manipulation of the proteins responsible for the amyloid disaggregation/degradation activities offers the possibility of ameliorating aggregation-associated diseases.
Figures



References
-
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. - PubMed
-
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. - PubMed
-
- Hammarstrom P, Schneider F, Kelly JW. Trans-suppression of misfolding in an amyloid disease. Science. 2001;293:2459–2462. - PubMed
-
- Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

