beta-Secretase cleavage is not required for generation of the intracellular C-terminal domain of the amyloid precursor family of proteins
- PMID: 20163459
- PMCID: PMC2847843
- DOI: 10.1111/j.1742-4658.2010.07579.x
beta-Secretase cleavage is not required for generation of the intracellular C-terminal domain of the amyloid precursor family of proteins
Abstract
The amyloid precursor family of proteins are of considerable interest, both because of their role in Alzheimer's disease pathogenesis and because of their normal physiological functions. In mammals, the amyloid precursor protein (APP) has two homologs, amyloid precursor-like protein (APLP) 1 and APLP2. All three proteins undergo ectodomain shedding and regulated intramembrane proteolysis, and important functions have been attributed to the full-length proteins, shed ectodomains, C-terminal fragments and intracellular domains (ICDs). One of the proteases that is known to cleave APP and that is essential for generation of the amyloid beta-protein is the beta-site APP-cleaving enzyme 1 (BACE1). Here, we investigated the effects of genetic manipulation of BACE1 on the processing of the APP family of proteins. BACE1 expression regulated the levels and species of full-length APLP1, APP and APLP2, of their shed ectodomains, and of their membrane-bound C-terminal fragments. In particular, APP processing appears to be tightly regulated, with changes in beta-cleaved APPs (APPsbeta) being compensated for by changes in alpha-cleaved APPs (APPsalpha). In contrast, the total levels of soluble cleaved APLP1 and APLP2 species were less tightly regulated, and fluctuated with BACE1 expression. Importantly, the production of ICDs for all three proteins was not decreased by loss of BACE1 activity. These results indicate that BACE1 is involved in regulating ectodomain shedding, maturation and trafficking of the APP family of proteins. Consequently, whereas inhibition of BACE1 is unlikely to adversely affect potential ICD-mediated signaling, it may alter other important facets of amyloid precursor-like protein/APP biology.
Figures
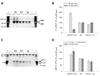
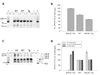
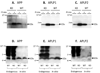
Similar articles
-
Determination of the proteolytic cleavage sites of the amyloid precursor-like protein 2 by the proteases ADAM10, BACE1 and γ-secretase.PLoS One. 2011;6(6):e21337. doi: 10.1371/journal.pone.0021337. Epub 2011 Jun 17. PLoS One. 2011. PMID: 21695060 Free PMC article.
-
Amyloid-β protein (Aβ) Glu11 is the major β-secretase site of β-site amyloid-β precursor protein-cleaving enzyme 1(BACE1), and shifting the cleavage site to Aβ Asp1 contributes to Alzheimer pathogenesis.Eur J Neurosci. 2013 Jun;37(12):1962-9. doi: 10.1111/ejn.12235. Eur J Neurosci. 2013. PMID: 23773065
-
Loss of cleavage at β'-site contributes to apparent increase in β-amyloid peptide (Aβ) secretion by β-secretase (BACE1)-glycosylphosphatidylinositol (GPI) processing of amyloid precursor protein.J Biol Chem. 2011 Jul 22;286(29):26166-77. doi: 10.1074/jbc.M111.260471. Epub 2011 Jun 3. J Biol Chem. 2011. PMID: 21642424 Free PMC article.
-
The beta-secretase, BACE: a prime drug target for Alzheimer's disease.J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
-
Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer's disease.Prog Neurobiol. 2006 Jun;79(2):95-111. doi: 10.1016/j.pneurobio.2006.06.001. Epub 2006 Aug 14. Prog Neurobiol. 2006. PMID: 16904810 Review.
Cited by
-
APLP1 is endoproteolytically cleaved by γ-secretase without previous ectodomain shedding.Sci Rep. 2018 Jan 30;8(1):1916. doi: 10.1038/s41598-018-19530-8. Sci Rep. 2018. PMID: 29382944 Free PMC article.
-
The Membrane-Bound Aspartyl Protease BACE1: Molecular and Functional Properties in Alzheimer's Disease and Beyond.Front Physiol. 2012 Feb 17;3:8. doi: 10.3389/fphys.2012.00008. eCollection 2012. Front Physiol. 2012. PMID: 22363289 Free PMC article.
-
Engineered Hsp70 chaperones prevent Aβ42-induced memory impairments in a Drosophila model of Alzheimer's disease.Sci Rep. 2018 Jul 2;8(1):9915. doi: 10.1038/s41598-018-28341-w. Sci Rep. 2018. PMID: 29967544 Free PMC article.
-
The transcriptionally active amyloid precursor protein (APP) intracellular domain is preferentially produced from the 695 isoform of APP in a {beta}-secretase-dependent pathway.J Biol Chem. 2010 Dec 31;285(53):41443-54. doi: 10.1074/jbc.M110.141390. Epub 2010 Oct 20. J Biol Chem. 2010. PMID: 20961856 Free PMC article.
-
Secreted amyloid-β precursor protein functions as a GABABR1a ligand to modulate synaptic transmission.Science. 2019 Jan 11;363(6423):eaao4827. doi: 10.1126/science.aao4827. Science. 2019. PMID: 30630900 Free PMC article.
References
-
- Selkoe DJ. Alzheimer's disease: genes, proteins, and therapy. Physiol Rev. 2001;81:741–766. - PubMed
-
- Coulson EJ, Paliga K, Beyreuther K, Masters CL. What the evolution of the amyloid protein precursor supergene family tells us about its function. Neurochem Int. 2000;36:175–184. - PubMed
-
- Wasco W, Gurubhagavatula S, Paradis MD, Romano DM, Sisodia SS, Hyman BT, Neve RL, Tanzi RE. Isolation and characterization of APLP2 encoding a homologue of the Alzheimer's associated amyloid beta protein precursor. Nat Genet. 1993;5:95–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

