Mitf-Mdel, a novel melanocyte/melanoma-specific isoform of microphthalmia-associated transcription factor-M, as a candidate biomarker for melanoma
- PMID: 20163701
- PMCID: PMC2839965
- DOI: 10.1186/1741-7015-8-14
Mitf-Mdel, a novel melanocyte/melanoma-specific isoform of microphthalmia-associated transcription factor-M, as a candidate biomarker for melanoma
Abstract
Background: Melanoma incidence is on the rise and advanced melanoma carries an extremely poor prognosis. Treatment options, including chemotherapy and immunotherapy, are limited and offer low response rates and transient efficacy. Thus, identification of new melanocyte/melanoma antigens that serve as potential novel candidate biomarkers in melanoma is an important area for investigation.
Methods: Full length MITF-M and its splice variant cDNA were cloned from human melanoma cell line 624 mel by reverse transcription polymerase chain reaction (RT-PCR). Expression was investigated using regular and quantitative RT-PCR in three normal melanocytes (NHEM), 31 melanoma cell lines, 21 frozen melanoma tissue samples, 18 blood samples (peripheral blood mononuclear cell; PBMC) from healthy donors and 12 non-melanoma cancer cell lines, including three breast, five glioma, one sarcoma, two kidney and one ovarian cancer cell lines.
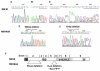
Results: A novel splice variant of MITF-M, which we named MITF-Mdel, was identified. The predicted MITF-Mdel protein contains two in frame deletions, 56- and 6- amino acid deletions in exon 2 (from V32 to E87) and exon 6 (from A187 to T192), respectively. MITF-Mdel was widely expressed in melanocytes, melanoma cell lines and tissues, but almost undetectable in non-melanoma cell lines or PBMC from healthy donors. Both isoforms were expressed significantly higher in melanoma tissues than in cell lines. Two of 31 melanoma cell lines expressed only one isoform or the other.
Conclusion: MITF-Mdel, a novel melanocyte/melanoma-specific isoform of MITF-M, may serve as a potential candidate biomarker for diagnostic and follow-up purposes in melanoma.
Figures



Similar articles
-
Expression of genes for microphthalmia isoforms, Pax3 and MSG1, in human melanomas.Cell Mol Biol (Noisy-le-grand). 1999 Nov;45(7):1075-82. Cell Mol Biol (Noisy-le-grand). 1999. PMID: 10644012
-
The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma.Cancer Res. 2002 Apr 1;62(7):2098-103. Cancer Res. 2002. PMID: 11929831
-
Microphthalmia transcription factor as a molecular marker for circulating tumor cell detection in blood of melanoma patients.Clin Cancer Res. 2006 Feb 15;12(4):1137-43. doi: 10.1158/1078-0432.CCR-05-1847. Clin Cancer Res. 2006. PMID: 16489066 Free PMC article.
-
[The Importance of MITF Signaling Pathway in the Regulation of Proliferation and Invasiveness of Malignant Melanoma].Klin Onkol. 2016 Fall;29(5):347-350. doi: 10.14735/amko2016347. Klin Onkol. 2016. PMID: 27739313 Review. Czech.
-
MITF: master regulator of melanocyte development and melanoma oncogene.Trends Mol Med. 2006 Sep;12(9):406-14. doi: 10.1016/j.molmed.2006.07.008. Epub 2006 Aug 8. Trends Mol Med. 2006. PMID: 16899407 Review.
Cited by
-
Activation of the long terminal repeat of human endogenous retrovirus K by melanoma-specific transcription factor MITF-M.Neoplasia. 2011 Nov;13(11):1081-92. doi: 10.1593/neo.11794. Neoplasia. 2011. PMID: 22131883 Free PMC article.
-
DUSP4 protects BRAF- and NRAS-mutant melanoma from oncogene overdose through modulation of MITF.Life Sci Alliance. 2022 May 17;5(9):e202101235. doi: 10.26508/lsa.202101235. Print 2022 Sep. Life Sci Alliance. 2022. PMID: 35580987 Free PMC article.
-
MITF in melanoma: mechanisms behind its expression and activity.Cell Mol Life Sci. 2015 Apr;72(7):1249-60. doi: 10.1007/s00018-014-1791-0. Epub 2014 Nov 30. Cell Mol Life Sci. 2015. PMID: 25433395 Free PMC article. Review.
-
The transcription factor MITF is a critical regulator of GPNMB expression in dendritic cells.Cell Commun Signal. 2015 Mar 24;13:19. doi: 10.1186/s12964-015-0099-5. Cell Commun Signal. 2015. PMID: 25889792 Free PMC article.
-
Evidence for an alternatively spliced MITF exon 2 variant.J Invest Dermatol. 2014 Apr;134(4):1166-1168. doi: 10.1038/jid.2013.426. Epub 2013 Oct 14. J Invest Dermatol. 2014. PMID: 24226203 No abstract available.
References
-
- Goding CR. Mitf from neural crest to melanoma: signal transduction and transcription in the melanocyte lineage. Genes Dev. 2000;14:1712–1728. - PubMed
-
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-T. - DOI - PubMed
-
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D'Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, Jenkins NA. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

