Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response
- PMID: 20164059
- PMCID: PMC2877989
- DOI: 10.1074/mcp.M900616-MCP200
Site-specific phosphorylation dynamics of the nuclear proteome during the DNA damage response
Abstract
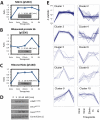
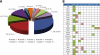
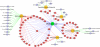
To investigate the temporal regulation of the DNA damage response, we applied quantitative mass spectrometry-based proteomics to measure site-specific phosphorylation changes of nuclear proteins after ionizing radiation. We profiled 5204 phosphorylation sites at five time points following DNA damage of which 594 sites on 209 proteins were observed to be regulated more than 2-fold. Of the 594 sites, 372 are novel phosphorylation sites primarily of nuclear origin. The 594 sites could be classified to distinct temporal profiles. Sites regulated shortly after radiation were enriched in the ataxia telangiectasia mutated (ATM) kinase SQ consensus sequence motif and a novel SXXQ motif. Importantly, in addition to induced phosphorylation, we identified a considerable group of sites that undergo DNA damage-induced dephosphorylation. Together, our data extend the number of known phosphorylation sites regulated by DNA damage, provides so far unprecedented temporal dissection of DNA damage-modified phosphorylation events, and elucidate the cross-talk between different types of post-translational modifications in the dynamic regulation of a multifaceted DNA damage response.
Figures


References
-
- Löbrich M., Jeggo P. A. (2007) The impact of a negligent G2/M checkpoint on genomic instability and cancer induction. Nat. Rev. Cancer 7, 861–869 - PubMed
-
- Bartek J., Lukas J. (2007) DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol 19, 238–245 - PubMed
-
- Zhou B. B., Elledge S. J. (2000) The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 - PubMed
-
- Lavin M. F. (2008) Ataxia-telangiectasia: from a rare disorder to a paradigm for cell signalling and cancer. Nat. Rev. Mol. Cell Biol 9, 759–769 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

