Minimizing off-target signals in RNA fluorescent in situ hybridization
- PMID: 20164092
- PMCID: PMC2879521
- DOI: 10.1093/nar/gkq042
Minimizing off-target signals in RNA fluorescent in situ hybridization
Abstract
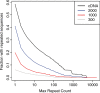
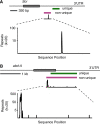
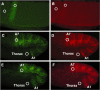
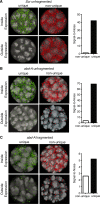
Fluorescent in situ hybridization (FISH) techniques are becoming extremely sensitive, to the point where individual RNA or DNA molecules can be detected with small probes. At this level of sensitivity, the elimination of 'off-target' hybridization is of crucial importance, but typical probes used for RNA and DNA FISH contain sequences repeated elsewhere in the genome. We find that very short (e.g. 20 nt) perfect repeated sequences within much longer probes (e.g. 350-1500 nt) can produce significant off-target signals. The extent of noise is surprising given the long length of the probes and the short length of non-specific regions. When we removed the small regions of repeated sequence from either short or long probes, we find that the signal-to-noise ratio is increased by orders of magnitude, putting us in a regime where fluorescent signals can be considered to be a quantitative measure of target transcript numbers. As the majority of genes in complex organisms contain repeated k-mers, we provide genome-wide annotations of k-mer-uniqueness at http://cbio.mskcc.org/ approximately aarvey/repeatmap.
Figures
References
-
- Kosman D, Mizutani C, Lemons D, Cox W, McGinnis W, Bier E. Multiplex detection of RNA expression in Drosophila embryos. Science. 2004;305:846. - PubMed
-
- Gregorieff A, Pinto D, Begthel H, Destrée O, Kielman M, Clevers H. Expression pattern of Wnt signaling components in the adult intestine. Gastroenterology. 2005;129:626–638. - PubMed
-
- Levsky J, Singer R. Fluorescence in situ hybridization: past, present and future. J. Cell Sci. 2003;116:2833–2838. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

