A two-step process controls the formation of the bienzyme cysteine synthase complex
- PMID: 20164178
- PMCID: PMC2857139
- DOI: 10.1074/jbc.M109.075762
A two-step process controls the formation of the bienzyme cysteine synthase complex
Abstract
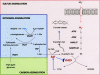
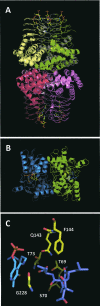
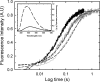
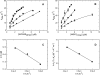
The regulation of enzyme activity through the transient formation of multiprotein assemblies plays an important role in the control of biosynthetic pathways. One of the first regulatory complexes to be discovered was cysteine synthase (CS), formed by the pyridoxal 5'-phosphate-dependent enzyme O-acetylserine sulfhydrylase (OASS) and serine acetyltransferase (SAT). These enzymes are at the branch point of the sulfur, carbon, and nitrogen assimilation pathways. Understanding the mechanism of complex formation helps to clarify the role played by CS in the regulation of sulfur assimilation in bacteria and plants. To this goal, stopped-flow fluorescence spectroscopy was used to characterize the interaction of SAT with OASS, at different temperatures and pH values, and in the presence of the physiological regulators cysteine and bisulfide. Results shed light on the mechanism of complex formation and regulation, so far poorly understood. Cysteine synthase assembly occurs via a two-step mechanism involving rapid formation of an encounter complex between the two enzymes, followed by a slow conformational change. The conformational change likely results from the closure of the active site of OASS upon binding of the SAT C-terminal peptide. Bisulfide, the second substrate and a feedback inhibitor of OASS, stabilizes the CS complex mainly by decreasing the back rate of the isomerization step. Cysteine, the product of the OASS reaction and a SAT inhibitor, slightly affects the kinetics of CS formation leading to destabilization of the complex.
Figures
References
-
- Ovadi J. (1991) J. Theor. Biol. 152, 1–22 - PubMed
-
- Huang X., Holden H. M., Raushel F. M. (2001) Annu. Rev. Biochem. 70, 149–180 - PubMed
-
- Miles E. W., Rhee S., Davies D. R. (1999) J. Biol. Chem. 274, 12193–12196 - PubMed
-
- Bernhard S. A. (1988) Cell Biophys. 12, 119–132 - PubMed
-
- Raboni S., Pioselli B., Bettati S., Mozzarelli A. (2003) Biochim. Biophys. Acta 1647, 157–160 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

