A novel protease-activated receptor-1 interactor, Bicaudal D1, regulates G protein signaling and internalization
- PMID: 20164183
- PMCID: PMC2857018
- DOI: 10.1074/jbc.M110.105403
A novel protease-activated receptor-1 interactor, Bicaudal D1, regulates G protein signaling and internalization
Abstract
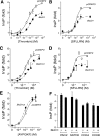
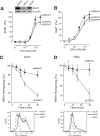
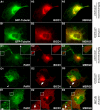
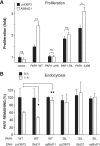
Protease-activated receptor-1 (PAR1) is a G protein-coupled receptor that plays critical roles in cancer, angiogenesis, inflammation, and thrombosis. Proteolytic cleavage of the extracellular domain of PAR1 generates a tethered ligand that activates PAR1 in an unusual intramolecular mode. The signal emanating from the irreversibly cleaved PAR1 is terminated by G protein uncoupling and internalization; however, the mechanisms of PAR1 signal shut off still remain unclear. Using a yeast two-hybrid screen, we identified Bicaudal D1 (BicD1) as a direct interactor with the C-terminal cytoplasmic domain of PAR1. BICD was originally identified as an essential developmental gene associated with mRNA and Golgi-endoplasmic reticulum transport. We discovered a novel function of BicD1 in the modulation of G protein signaling, cell proliferation, and endocytosis downstream of PAR1. BicD1 and its C-terminal CC3 domain inhibited PAR1 signaling to G(q)-phospholipase C-beta through coiled-coil interactions with the cytoplasmic 8th helix of PAR1. Unexpectedly, BicD1 was also found to be a potent suppressor of PAR1-driven proliferation of breast carcinoma cells. The growth-suppressing effects of BicD1 required the ability to interact with the 8th helix of PAR1. Silencing of BicD1 expression impaired endocytosis of PAR1, and BicD1 co-localized with PAR1 and tubulin, implicating BicD1 as an important adapter protein involved in the transport of PAR1 from the plasma membrane to endosomal vesicles. Together, these findings provide a link between PAR1 signal termination and internalization through the non-G protein effector, BicD1.
Figures


References
-
- Ossovskaya V. S., Bunnett N. W. (2004) Physiol. Rev. 84, 579–621 - PubMed
-
- Vu T.-K. H., Hung D. T., Wheaton V. I., Coughlin S. R. (1991) Cell 64, 1057–1068 - PubMed
-
- Kuliopulos A., Covic L., Seeley S. K., Sheridan P. J., Helin J., Costello C. E. (1999) Biochemistry 38, 4572–4585 - PubMed
-
- Riewald M., Petrovan R. J., Donner A., Mueller B. M., Ruf W. (2002) Science 296, 1880–1882 - PubMed
-
- Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. (2005) Cell 120, 303–313 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

