Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display
- PMID: 20164216
- PMCID: PMC2841545
- DOI: 10.1093/protein/gzq003
Generating a panel of highly specific antibodies to 20 human SH2 domains by phage display
Abstract
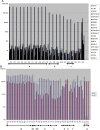
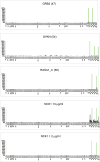
To demonstrate the utility of phage display in generating highly specific antibodies, affinity selections were conducted on 20 related Src Homology 2 (SH2) domains (ABL1, ABL2, BTK, BCAR3, CRK, FYN, GRB2, GRAP2, LYN, LCK, NCK1, PTPN11 C, PIK3R1 C, PLCgamma1 C, RASA1 C, SHC1, SH2D1A, SYK N, VAV1 and the tandem domains of ZAP70). The domains were expressed in Escherichia coli, purified and used in affinity selection experiments. In total, 1292/3800 of the resultant antibodies were shown to bind the target antigen. Of the 695 further evaluated in specificity ELISAs against all 20 SH2 domains, 379 antibodies were identified with unique specificity (i.e. monospecific). Sequence analysis revealed that there were at least 150 different clones with 1-19 different antibodies/antigen. This includes antibodies that distinguish between ABL1 and ABL2, despite their 89% sequence identity. Specificity was confirmed for many on protein arrays fabricated with 432 different proteins. Thus, even though the SH2 domains share a common three-dimensional structure and 20-89% identity at the primary structure level, we were able to isolate antibodies with exquisite specificity within this family of structurally related domains.
Figures



References
-
- Editorial. Nat. Methods. 2008;5:851.
-
- Haab B.B., Paulovich A.G., Anderson N.L., Clark A.M., Downing G.J., Hermjakob H., Labaer J., Uhlen M. Mol. Cell. Proteomics. 2006;5:1996–2007. - PubMed
-
- Huang H., et al. Mol. Cell. Proteomics. 2008;7:768–784. - PubMed
-
- Huber T., Steiner D., Rothlisberger D., Pluckthun A. J. Struct. Biol. 2007;159:206–221. - PubMed
-
- Kasai T., Koshiba S., Inoue M., Kigawa T., Yokoyama S. doi:10.2210/pdb2ecd/pdb. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

