Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes
- PMID: 20164218
- PMCID: PMC2863730
- DOI: 10.1128/JVI.00851-09
Human T-lymphotropic virus type 1 transcription and chromatin-remodeling complexes
Abstract
Human T-lymphotropic virus type 1 (HTLV-1) encodes the viral protein Tax, which is believed to act as a viral transactivator through its interactions with a variety of transcription factors, including CREB and NF-kappaB. As is the case for all retroviruses, the provirus is inserted into the host DNA, where nucleosomes are deposited to ensure efficient packaging. Nucleosomes act as roadblocks in transcription, making it difficult for RNA polymerase II (Pol II) to proceed toward the 3' end of the genome. Because of this, a variety of chromatin remodelers can act to modify nucleosomes, allowing for efficient transcription. While a number of covalent modifications are known to occur on histone tails in HTLV-1 infection (i.e., histone acetyltransferases [HATs], histone deacetylases [HDACs], and histone methyltransferases [HMTs]), evidence points to the use of chromatin remodelers that use energy from ATP hydrolysis to remodel nucleosomes. Here we confirm that BRG1, which is the core subunit of eight chromatin-remodeling complexes, is essential not only for Tax transactivation but also for viral replication. This is especially evident when wild-type infectious clones of HTLV-1 are used. BRG1 associates with Tax at the HTLV-1 long terminal repeat (LTR), and coexpression of BRG1 and Tax results in increased rates of transcription. The interaction of BRG1 with Tax additionally recruits the basal transcriptional machinery and removes some of the core histones from the nucleosome at the start site (Nuc 1). When using the BRG1-deficient cell lines SW13, C33A, and TSUPR1, we observed little viral transcription and no viral replication. Importantly, while these three cell lines do not express detectable levels of BRG1, much of the SWI/SNF complex remains assembled in the cells. Knockdown of BRG1 and associated SWI/SNF subunits suggests that the BRG1-utilizing SWI/SNF complex PBAF is responsible for HTLV-1 nucleosome remodeling. Finally, HTLV-1 infection of cell lines with a knockdown in BRG1 or the PBAF complex results in a significant reduction in viral production. Overall, we concluded that BRG1 is required for Tax transactivation and HTLV-1 viral production and that the PBAF complex appears to be responsible for nucleosome remodeling.
Figures

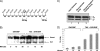




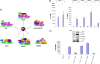
References
-
- Akagi, T., H. Ono, and K. Shimotohno. 1996. Expression of cell-cycle regulatory genes in HTLV-I infected T-cell lines: possible involvement of Tax1 in the altered expression of cyclin D2, p18Ink4 and p21Waf1/Cip1/Sdi1. Oncogene 12:1645-1652. - PubMed
-
- Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. - PubMed
-
- Brower-Toland, B., D. A. Wacker, R. M. Fulbright, J. T. Lis, W. L. Kraus, and M. D. Wang. 2005. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346:135-146. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

