Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain
- PMID: 20164234
- PMCID: PMC2863754
- DOI: 10.1128/JVI.02619-09
Two N-linked glycosylation sites in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 regulate viral neutralization susceptibility to the human monoclonal antibody specific for the CD4 binding domain
Abstract
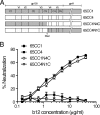
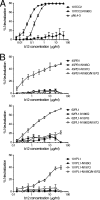
A recombinant human monoclonal antibody, IgG1 b12 (b12), recognizes a conformational epitope on human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) gp120 that overlaps the CD4 binding domain. Although b12 is able to broadly neutralize HIV-1 subtype B, C, and D viruses, many HIV-1 CRF01_AE viruses are resistant to b12-mediated neutralization. In this report, we examined the molecular mechanisms underlying the low neutralization susceptibility of CRF01_AE viruses to b12, using recently established CRF01_AE Env recombinant viruses. Our results showed that two potential N-linked glycosylation (PNLG) sites in the V2 and C2 regions of Env gp120 played an important role in regulating the susceptibility of CRF01_AE Env to b12. The locations of these PNLG sites correspond to amino acid positions 186 and 197 in HXB2 Env gp120; thus, they are designated N186 and N197 in this study. Removal of N186 significantly conferred the b12 susceptibility of 2 resistant CRF01_AE Env clones, 65CC4 and 107CC2, while the introduction of N186 reduced the b12 susceptibility of a susceptible CRF01_AE Env clone, 65CC1. In addition, removal of both N186 and N197 conferred the b12 susceptibility of 3 resistant CRF01_AE Env clones, 45PB1, 62PL1, and 101PL1, whereas the removal of either N186 or N197 was not sufficient to confer the b12 susceptibility of these CRF01_AE Env clones. Finally, removal of N197 conferred the b12 susceptibility of 2 resistant CRF01_AE Env clones lacking N186, 55PL1 and 102CC2. Taken together, we propose that two PNLG sites, N186 and N197, in Env gp120 are important determinants of the b12 resistance of CRF01_AE viruses.
Figures



Similar articles
-
Three amino acid residues in the envelope of human immunodeficiency virus type 1 CRF07_BC regulate viral neutralization susceptibility to the human monoclonal neutralizing antibody IgG1b12.Virol Sin. 2014 Oct;29(5):299-307. doi: 10.1007/s12250-014-3485-z. Epub 2014 Sep 28. Virol Sin. 2014. PMID: 25273335 Free PMC article.
-
Impact of amino acid substitutions in the V2 and C2 regions of human immunodeficiency virus type 1 CRF01_AE envelope glycoprotein gp120 on viral neutralization susceptibility to broadly neutralizing antibodies specific for the CD4 binding site.Retrovirology. 2014 Apr 23;11:32. doi: 10.1186/1742-4690-11-32. Retrovirology. 2014. PMID: 24758333 Free PMC article.
-
A 2-4-Amino Acid Deletion in the V5 Region of HIV-1 Env gp120 Confers Viral Resistance to the Broadly Neutralizing Human Monoclonal Antibody, VRC01.AIDS Res Hum Retroviruses. 2017 Dec;33(12):1248-1257. doi: 10.1089/aid.2017.0063. Epub 2017 Oct 17. AIDS Res Hum Retroviruses. 2017. PMID: 28903577
-
Phenotypic studies on recombinant human immunodeficiency virus type 1 (HIV-1) containing CRF01_AE env gene derived from HIV-1-infected patient, residing in central Thailand.Microbes Infect. 2009 Mar;11(3):334-43. doi: 10.1016/j.micinf.2008.12.008. Epub 2008 Dec 27. Microbes Infect. 2009. PMID: 19136072
-
Glycans flanking the hypervariable connecting peptide between the A and B strands of the V1/V2 domain of HIV-1 gp120 confer resistance to antibodies that neutralize CRF01_AE viruses.PLoS One. 2015 Mar 20;10(3):e0119608. doi: 10.1371/journal.pone.0119608. eCollection 2015. PLoS One. 2015. PMID: 25793890 Free PMC article.
Cited by
-
Changes in Structure and Antigenicity of HIV-1 Env Trimers Resulting from Removal of a Conserved CD4 Binding Site-Proximal Glycan.J Virol. 2016 Sep 29;90(20):9224-36. doi: 10.1128/JVI.01116-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27489265 Free PMC article.
-
N463 Glycosylation Site on V5 Loop of a Mutant gp120 Regulates the Sensitivity of HIV-1 to Neutralizing Monoclonal Antibodies VRC01/03.J Acquir Immune Defic Syndr. 2015 Jul 1;69(3):270-7. doi: 10.1097/QAI.0000000000000595. J Acquir Immune Defic Syndr. 2015. PMID: 25751231 Free PMC article.
-
The selection of low envelope glycoprotein reactivity to soluble CD4 and cold during simian-human immunodeficiency virus infection of rhesus macaques.J Virol. 2014 Jan;88(1):21-40. doi: 10.1128/JVI.01558-13. Epub 2013 Oct 16. J Virol. 2014. PMID: 24131720 Free PMC article.
-
Three amino acid residues in the envelope of human immunodeficiency virus type 1 CRF07_BC regulate viral neutralization susceptibility to the human monoclonal neutralizing antibody IgG1b12.Virol Sin. 2014 Oct;29(5):299-307. doi: 10.1007/s12250-014-3485-z. Epub 2014 Sep 28. Virol Sin. 2014. PMID: 25273335 Free PMC article.
-
HIV-1 subtype CRF01_AE and B differ in utilization of low levels of CCR5, Maraviroc susceptibility and potential N-glycosylation sites.Virology. 2017 Dec;512:222-233. doi: 10.1016/j.virol.2017.09.026. Epub 2017 Oct 9. Virology. 2017. PMID: 29020646 Free PMC article.
References
-
- Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. - PubMed
-
- Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. - PMC - PubMed
-
- Burton, D. R., C. F. Barbas III, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 88:10134-10137. - PMC - PubMed
-
- Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

