Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses
- PMID: 20164239
- PMCID: PMC2940668
- DOI: 10.1093/neuonc/nop075
Distinct patterns of 1p and 19q alterations identify subtypes of human gliomas that have different prognoses
Abstract
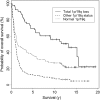
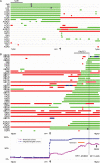
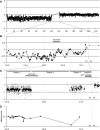
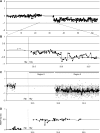
We studied the status of chromosomes 1 and 19 in 363 astrocytic and oligodendroglial tumors. Whereas the predominant pattern of copy number abnormality was a concurrent loss of the entire 1p and 19q regions (total 1p/19q loss) among oligodendroglial tumors and partial deletions of 1p and/or 19q in astrocytic tumors, a subset of apparently astrocytic tumors also had total 1p/19q loss. The presence of total 1p/19q loss was associated with longer survival of patients with all types of adult gliomas independent of age and diagnosis (P = .041). The most commonly deleted region on 19q in astrocytic tumors spans 885 kb in 19q13.33-q13.41, which is telomeric to the previously proposed region. Novel regions of homozygous deletion, including a part of DPYD (1p21.3) or the KLK cluster (19q13.33), were observed in anaplastic oligodendrogliomas. Amplifications encompassing AKT2 (19q13.2) or CCNE1 (19q12) were identified in some glioblastomas. Deletion mapping of the centromeric regions of 1p and 19q in the tumors that had total 1p/19q loss, indicating that the breakpoints lie centromeric to NOTCH2 within the pericentromeric regions of 1p and 19q. Thus, we show that the copy number abnormalities of 1p and 19q in human gliomas are complex and have distinct patterns that are prognostically predictive independent of age and pathological diagnosis. An accurate identification of total 1p/19q loss and discriminating this from other 1p/19q changes is, however, critical when the 1p/19q copy number status is used to stratify patients in clinical trials.
Figures

References
-
- Griffin CA, Burger P, Morsberger L, et al. Identification of der(1;19)(q10;p10) in five oligodendrogliomas suggests mechanism of concurrent 1p and 19q loss. J Neuropathol Exp Neurol. 2006;65:988–994. - PubMed
-
- Jenkins RB, Blair H, Ballman KV, et al. A t(1;19)(q10;p10) mediates the combined deletions of 1p and 19q and predicts a better prognosis of patients with oligodendroglioma. Cancer Res. 2006;66:9852–9861. - PubMed
-
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK. WHO Classification of Tumours of the Central Nervous System. 4th. Lyon, France: International Agency for Research on Cancer; 2007.
-
- Cairncross G, Berkey B, Shaw E, et al. Phase III trial of chemotherapy plus radiotherapy compared with radiotherapy alone for pure and mixed anaplastic oligodendroglioma: Intergroup Radiation Therapy Oncology Group Trial 9402. J Clin Oncol. 2006;24:2707–2714. - PubMed
-
- van den Bent MJ, Carpentier AF, Brandes AA, et al. Adjuvant procarbazine, lomustine, and vincristine improves progression-free survival but not overall survival in newly diagnosed anaplastic oligodendrogliomas and oligoastrocytomas: a randomized European Organisation for Research and Treatment of Cancer phase III trial. J Clin Oncol. 2006;24:2715–2722. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

