Regulation of the subcellular distribution of key cellular RNA-processing factors during permissive human cytomegalovirus infection
- PMID: 20164265
- PMCID: PMC2888166
- DOI: 10.1099/vir.0.020313-0
Regulation of the subcellular distribution of key cellular RNA-processing factors during permissive human cytomegalovirus infection
Abstract
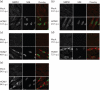
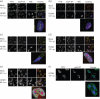
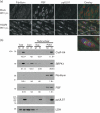
Alternative splicing and polyadenylation of human cytomegalovirus (HCMV) immediate-early (IE) pre-mRNAs are temporally regulated and rely on cellular RNA-processing factors. This study examined the location and abundance of essential RNA-processing factors, which affect alternative processing of UL37 IE pre-mRNAs, during HCMV infection. Serine/threonine protein kinase 1 (SRPK1) phosphorylates serine/arginine-rich proteins, necessary for pre-spliceosome commitment. It was found that HCMV infection progressively increased the abundance of cytoplasmic SRPK1, which is regulated by subcellular partitioning. The essential polyadenylation factor CstF-64 was similarly increased in abundance, albeit in the nucleus, proximal to and within viral replication compartments (VRCs). In contrast, the location of polypyrimidine tract-binding protein (PTB), known to adversely affect splicing of HCMV major IE RNAs, was temporally regulated during infection. PTB co-localized with CstF-64 in the nucleus at IE times. By early times, PTB was detected in punctate cytoplasmic sites of some infected cells. At late times, PTB relocalized to the nucleus, where it was notably excluded from HCMV VRCs. Moreover, HCMV infection induced the formation of nucleolar stress structures, fibrillarin-containing caps, in close proximity to its VRCs. PTB exclusion from HCMV VRCs required HCMV DNA synthesis and/or late gene expression, whereas the regulation of SRPK1 subcellular distribution did not. Taken together, these results indicated that HCMV increasingly regulates the subcellular distribution and abundance of essential RNA-processing factors, thereby altering their ability to affect the processing of viral pre-mRNAs. These results further suggest that HCMV infection selectively induces sorting of nucleolar and nucleoplasmic components.
Figures




Similar articles
-
Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection.J Gen Virol. 2004 Dec;85(Pt 12):3541-3553. doi: 10.1099/vir.0.80450-0. J Gen Virol. 2004. PMID: 15557227
-
Roles of polypyrimidine tract binding proteins in major immediate-early gene expression and viral replication of human cytomegalovirus.J Virol. 2009 Apr;83(7):2839-50. doi: 10.1128/JVI.02407-08. Epub 2009 Jan 14. J Virol. 2009. PMID: 19144709 Free PMC article.
-
Convergence of RNA cis elements and cellular polyadenylation factors in the regulation of human cytomegalovirus UL37 exon 1 unspliced RNA production.J Virol. 2003 Dec;77(23):12729-41. doi: 10.1128/jvi.77.23.12729-12741.2003. J Virol. 2003. PMID: 14610195 Free PMC article.
-
[Interrelationship between human cytomegalovirus infection and chemokine].Nihon Rinsho. 1998 Jan;56(1):69-74. Nihon Rinsho. 1998. PMID: 9465667 Review. Japanese.
-
Early viral gene expression and function.In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. Chapter 18. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K, editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. Chapter 18. PMID: 21348120 Free Books & Documents. Review.
Cited by
-
Two Polypyrimidine Tracts in Intron 4 of the Major Immediate Early Gene Are Critical for Gene Expression Switching from IE1 to IE2 and for Replication of Human Cytomegalovirus.J Virol. 2016 Jul 27;90(16):7339-7349. doi: 10.1128/JVI.00837-16. Print 2016 Aug 15. J Virol. 2016. PMID: 27252533 Free PMC article.
-
Host proviral and antiviral factors for SARS-CoV-2.Virus Genes. 2021 Dec;57(6):475-488. doi: 10.1007/s11262-021-01869-2. Epub 2021 Sep 11. Virus Genes. 2021. PMID: 34510331 Free PMC article. Review.
-
Interplay Between CMGC Kinases Targeting SR Proteins and Viral Replication: Splicing and Beyond.Front Microbiol. 2021 Mar 29;12:658721. doi: 10.3389/fmicb.2021.658721. eCollection 2021. Front Microbiol. 2021. PMID: 33854493 Free PMC article. Review.
-
Host protein kinases required for SARS-CoV-2 nucleocapsid phosphorylation and viral replication.Sci Signal. 2022 Oct 25;15(757):eabm0808. doi: 10.1126/scisignal.abm0808. Epub 2022 Oct 25. Sci Signal. 2022. PMID: 36282911 Free PMC article.
-
The structural basis of CstF-77 modulation of cleavage and polyadenylation through stimulation of CstF-64 activity.Nucleic Acids Res. 2018 Dec 14;46(22):12022-12039. doi: 10.1093/nar/gky862. Nucleic Acids Res. 2018. PMID: 30257008 Free PMC article.
References
-
- Adair, R., Liebisch, G. W. & Colberg-Poley, A. M. (2003). Complex alternative processing of human cytomegalovirus UL37 pre-mRNA. J Gen Virol 84, 3353–3358. - PubMed
-
- Adair, R., Liebisch, G. W., Su, Y. & Colberg-Poley, A. M. (2004). Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J Gen Virol 85, 3541–3553. - PubMed
-
- Adair, R., Liebisch, G. W., Lerman, B. J. & Colberg-Poley, A. M. (2006). Human cytomegalovirus temporally regulated gene expression in differentiated, immortalized retinal pigment epithelial cells. J Clin Virol 35, 478–484. - PubMed
-
- Andersen, J. S., Lyon, C. E., Fox, A. H., Leung, A. K., Lam, Y. W., Steen, H., Mann, M. & Lamond, A. I. (2002). Directed proteomic analysis of the human nucleolus. Curr Biol 12, 1–11. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

