Development of real-time PCR assays for detection of the Streptococcus milleri group from cystic fibrosis clinical specimens by targeting the cpn60 and 16S rRNA genes
- PMID: 20164275
- PMCID: PMC2849594
- DOI: 10.1128/JCM.02082-09
Development of real-time PCR assays for detection of the Streptococcus milleri group from cystic fibrosis clinical specimens by targeting the cpn60 and 16S rRNA genes
Abstract
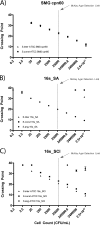
Cystic fibrosis (CF) is a multiorgan disease, with the majority of mortalities resulting from pulmonary failure due to repeated pulmonary exacerbations. Recently, members of the Streptococcus anginosus group (S. anginosus, S. constellatus, and S. intermedius), herein referred to as the "Streptococcus milleri group" (SMG) have been implicated as important etiological pathogens contributing to pulmonary exacerbations in CF patients. This is partly due to better microbiological detection of the SMG species through the development of a novel specific medium termed "McKay agar." McKay agar demonstrated that SMG has been an underreported respiratory pathogen contributing to lung exacerbations. Our aim was to develop a real-time PCR assay to expedite the detection of SMG within diagnostic samples. The cpn60 gene was chosen as a target, with all three members amplified using a single hybridization probe set. SMG strain analysis showed that speciation based on melting curve analysis allowed for the majority of the S. constellatus (96%), S. intermedius (94%), and S. anginosus (60%) strains to be correctly identified. To increase specificity for S. anginosus, two 16S rRNA real-time PCR assays were developed targeting the 16S rRNA gene. The 16s_SA assay is specific for S. anginosus (100%), while the 16s_SCI assay is specific for S. constellatus and S. intermedius (100%). These assays can detect <10 genome equivalents in pure culture and >10(4) genome equivalents in sputum samples, making this a great tool for assessment of the presence of SMG in complex polymicrobial samples. Novel molecular methods were developed providing detection ability for SMG, an emerging opportunistic pathogen.
Figures

Similar articles
-
McKay agar enables routine quantification of the 'Streptococcus milleri' group in cystic fibrosis patients.J Med Microbiol. 2010 May;59(Pt 5):534-540. doi: 10.1099/jmm.0.016592-0. Epub 2010 Jan 21. J Med Microbiol. 2010. PMID: 20093379
-
Evaluation of genotypic and phenotypic methods for differentiation of the members of the Anginosus group streptococci.Eur J Clin Microbiol Infect Dis. 2009 Sep;28(9):1123-8. doi: 10.1007/s10096-009-0758-9. Epub 2009 Jun 4. Eur J Clin Microbiol Infect Dis. 2009. PMID: 19495818 Free PMC article.
-
Characterization of Streptococcus milleri group isolates from expectorated sputum of adult patients with cystic fibrosis.J Clin Microbiol. 2010 Feb;48(2):395-401. doi: 10.1128/JCM.01807-09. Epub 2009 Dec 9. J Clin Microbiol. 2010. PMID: 20007382 Free PMC article.
-
Molecular pathogenicity of Streptococcus anginosus.Mol Oral Microbiol. 2014 Aug;29(4):145-55. doi: 10.1111/omi.12056. Epub 2014 Jun 26. Mol Oral Microbiol. 2014. PMID: 24848553 Review.
-
Streptococcus intermedius causing infective endocarditis and abscesses: a report of three cases and review of the literature.BMC Infect Dis. 2008 Nov 10;8:154. doi: 10.1186/1471-2334-8-154. BMC Infect Dis. 2008. PMID: 18992173 Free PMC article. Review.
Cited by
-
The Streptococcus milleri population of a cystic fibrosis clinic reveals patient specificity and intraspecies diversity.J Clin Microbiol. 2010 Jul;48(7):2592-4. doi: 10.1128/JCM.00414-10. Epub 2010 May 12. J Clin Microbiol. 2010. PMID: 20463160 Free PMC article.
-
A New Highly Sensitive and Specific Real-Time PCR Assay Targeting the Malate Dehydrogenase Gene of Kingella kingae and Application to 201 Pediatric Clinical Specimens.J Clin Microbiol. 2018 Jul 26;56(8):e00505-18. doi: 10.1128/JCM.00505-18. Print 2018 Aug. J Clin Microbiol. 2018. PMID: 29875189 Free PMC article.
-
Streptococcus intermedius: an underestimated pathogen in brain infection?Future Microbiol. 2025 Feb;20(2):163-177. doi: 10.1080/17460913.2024.2423524. Epub 2024 Nov 18. Future Microbiol. 2025. PMID: 39552595 Free PMC article. Review.
-
Direct sampling of cystic fibrosis lungs indicates that DNA-based analyses of upper-airway specimens can misrepresent lung microbiota.Proc Natl Acad Sci U S A. 2012 Aug 21;109(34):13769-74. doi: 10.1073/pnas.1107435109. Epub 2012 Aug 7. Proc Natl Acad Sci U S A. 2012. PMID: 22872870 Free PMC article.
-
Auto-aggregation in Streptococcus intermedius is driven by the Pel polysaccharide.mBio. 2025 Aug 13;16(8):e0119625. doi: 10.1128/mbio.01196-25. Epub 2025 Jul 7. mBio. 2025. PMID: 40621902 Free PMC article.
References
-
- Buckley, D. A., A. Murphy, P. Dervan, R. Hone, T. O'Dwyer, and S. O'Loughlin. 1998. Persistent infection of the chin with an unusual skin pathogen (Streptococcus milleri): a sign of intraoral carcinoma. Clin. Exp. Dermatol. 23:35-37. - PubMed
-
- Chen, H. J., J. C. Tsai, T. C. Chang, W. C. Hung, S. P. Tseng, P. R. Hsueh, and L. J. Teng. 2008. PCR-RFLP assay for species and subspecies differentiation of the Streptococcus bovis group based on groESL sequences. J. Med. Microbiol. 57:432-438. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

