Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles
- PMID: 20164450
- PMCID: PMC2904001
- DOI: 10.1167/iovs.09-4660
Somatic mitochondrial DNA deletions accumulate to high levels in aging human extraocular muscles
Abstract
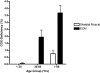
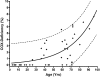
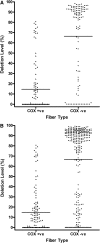
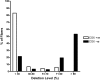
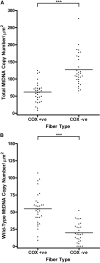
PURPOSE. Mitochondrial function and the presence of somatic mitochondrial DNA (mtDNA) defects were investigated in extraocular muscles (EOMs) collected from individuals covering a wide age range, to document the changes seen with normal aging. METHODS. Cytochrome c oxidase (COX) and succinate dehydrogenase (SDH) histochemistry was performed on 46 EOM samples to determine the level of COX deficiency in serial cryostat muscle sections (mean age, 42.6 years; range, 3.0-96.0 years). Competitive three-primer and real-time PCR were performed on single-fiber lysates to detect and quantify mtDNA deletions. Whole-genome mitochondrial sequencing was also performed to evaluate the contribution of mtDNA point mutations to the overall mutational load. RESULTS. COX-negative fibers were seen in EOMs beginning in the third decade of life, and there was a significant age-related increase: <30 years, 0.05% (n = 17); 30 to 60 years, 1.94% (n = 13); and >60 years, 3.34% (n = 16, P = 0.0001). Higher levels of COX deficiency were also present in EOM than in skeletal muscle in all three age groups (P < 0.0001). Most of the COX-negative fibers harbored high levels (>70%) of mtDNA deletions (206/284, 72.54%) and the mean deletion level was 66.64% (SD 36.45%). The mutational yield from whole mitochondrial genome sequencing was relatively low (1/19, 5.3%), with only a single mtDNA point mutation identified among COX-negative fibers with low deletion levels < or =70%. CONCLUSIONS. The results show an exponential increase in COX deficiency in EOMs beginning in early adulthood, which suggests an accelerated aging process compared with other postmitotic tissues.
Figures
References
-
- Schapira AHV. Mitochondrial disease. Lancet 2006;368:70–82 - PubMed
-
- Schaefer AM, McFarland R, Blakely EL, et al. Prevalence of mitochondrial DNA disease in adults. Ann Neurol 2008;63:35–39 - PubMed
-
- Manwaring N, Jones MM, Wang JJ, et al. Population prevalence of the MELAS A3243G mutation. Mitochondrion 2007;7:230–233 - PubMed
-
- Shoubridge EA, Karpati G, Hastings KEM. Deletion mutants are functionally dominant over wild-type mitochondrial genomes in skeletal-muscle fiber segments in mitochondrial disease. Cell 1990;62:43–49 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

