Spectrum of rhodopsin mutations in French autosomal dominant rod-cone dystrophy patients
- PMID: 20164459
- PMCID: PMC3102265
- DOI: 10.1167/iovs.09-4766
Spectrum of rhodopsin mutations in French autosomal dominant rod-cone dystrophy patients
Abstract
PURPOSE. To identify the prevalence of rhodopsin (RHO) mutations in French patients with autosomal dominant rod-cone dystrophies (adRPs). Methods. Detailed phenotypic characterization was performed, including precise family history, best corrected visual acuity with the ETDRS chart, slit lamp examination, kinetic and static perimetry, full-field and multifocal electroretinography (ERG), fundus autofluorescence imaging (FAF), and optical coherence tomography (OCT). For genetic diagnosis, genomic DNA of 79 families was isolated by standard
Methods: The coding exons and flanking intronic regions of RHO were PCR amplified, purified, and sequenced in the index patient. RESULTS. Of this French adRP sample, 16.5% carried an RHO mutation. Three different families showed a novel mutation (p. Leu88Pro, p.Met207Lys and p.Gln344Pro), while ten unrelated families showed recurrent, previously published mutations (p.Asn15Ser, p.Leu131Pro, p.Arg135Trp, p.Ser334GlyfsX21 and p.Pro347Leu). All mutations co-segregated with the phenotype within a family, and the novel mutations were not identified in control samples. CONCLUSIONS. This study revealed that the prevalence of RHO mutations in French adRP patients is in accordance with that in other studies from Europe. Most of the changes identified herein reflect recurrent mutations, within which p.Pro347Leu substitution is the most prevalent. Nevertheless, almost one fourth of the changes are novel, indicating that, although RHO is the first gene implicated and probably the most studied gene in RP, it is still important performing mutation analysis in RHO to detect novel changes. The detailed phenotype-genotype analyses in all available family members deliver the basis for therapeutic approaches in those families.
Figures
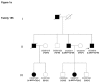













References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

