In front of and behind the replication fork: bacterial type IIA topoisomerases
- PMID: 20165898
- PMCID: PMC11115839
- DOI: 10.1007/s00018-010-0299-5
In front of and behind the replication fork: bacterial type IIA topoisomerases
Abstract
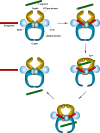
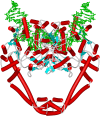
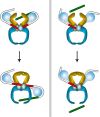
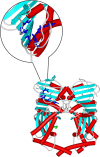
Topoisomerases are vital enzymes specialized in controlling DNA topology, in particular supercoiling and decatenation, to properly handle nucleic acid packing and cell dynamics. The type IIA enzymes act by cleaving both strands of a double helix and having another strand from the same or another molecule cross the DNA gate before a re-sealing event completes the catalytic cycle. Here, we will consider the two types of IIA prokaryotic topoisomerases, DNA Gyrase and Topoisomerase IV, as crucial regulators of bacterial cell cycle progression. Their synergistic action allows control of chromosome packing and grants occurrence of functional transcription and replication processes. In addition to displaying a fascinating molecular mechanism of action, which transduces chemical energy into mechanical energy by means of large conformational changes, these enzymes represent attractive pharmacological targets for antibacterial chemotherapy.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

