DomSVR: domain boundary prediction with support vector regression from sequence information alone
- PMID: 20165918
- PMCID: PMC2909371
- DOI: 10.1007/s00726-010-0506-6
DomSVR: domain boundary prediction with support vector regression from sequence information alone
Abstract
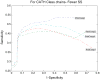
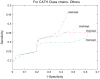
Protein domains are structural and fundamental functional units of proteins. The information of protein domain boundaries is helpful in understanding the evolution, structures and functions of proteins, and also plays an important role in protein classification. In this paper, we propose a support vector regression-based method to address the problem of protein domain boundary identification based on novel input profiles extracted from AAindex database. As a result, our method achieves an average sensitivity of approximately 36.5% and an average specificity of approximately 81% for multi-domain protein chains, which is overall better than the performance of published approaches to identify domain boundary. As our method used sequence information alone, our method is simpler and faster.
Figures
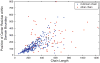


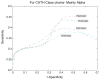
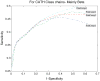
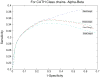


References
-
- Baldi P, Brunak S, Chauvin Y, Andersen CA, Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. - PubMed
-
- Chen P, Wang B, Wong HS, Huang DS. Prediction of protein B-factors using multi-class bounded SVM. Protein Pept Lett. 2007;14(2):185–190. - PubMed
-
- Cheng J, Sweredoski MJ, Baldi P. DOMpro: protein domain prediction using profiles, secondary structure, relative solvent accessibility, and recursive neural networks. Data Min Knowl Discov. 2006;13:1–10.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

