Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA
- PMID: 20167078
- PMCID: PMC2834691
- DOI: 10.1186/1471-2148-10-50
Mitochondrial phylogenomics of the Bivalvia (Mollusca): searching for the origin and mitogenomic correlates of doubly uniparental inheritance of mtDNA
Abstract
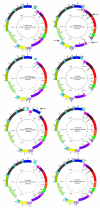
Background: Doubly uniparental inheritance (DUI) is an atypical system of animal mtDNA inheritance found only in some bivalves. Under DUI, maternally (F genome) and paternally (M genome) transmitted mtDNAs yield two distinct gender-associated mtDNA lineages. The oldest distinct M and F genomes are found in freshwater mussels (order Unionoida). Comparative analyses of unionoid mitochondrial genomes and a robust phylogenetic framework are necessary to elucidate the origin, function and molecular evolutionary consequences of DUI. Herein, F and M genomes from three unionoid species, Venustaconcha ellipsiformis, Pyganodon grandis and Quadrula quadrula have been sequenced. Comparative genomic analyses were carried out on these six genomes along with two F and one M unionoid genomes from GenBank (F and M genomes of Inversidens japanensis and F genome of Lampsilis ornata).
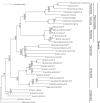
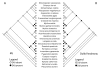
Results: Compared to their unionoid F counterparts, the M genomes contain some unique features including a novel localization of the trnH gene, an inversion of the atp8-trnD genes and a unique 3'coding extension of the cytochrome c oxidase subunit II gene. One or more of these unique M genome features could be causally associated with paternal transmission. Unionoid bivalves are characterized by extreme intraspecific sequence divergences between gender-associated mtDNAs with an average of 50% for V. ellipsiformis, 50% for I. japanensis, 51% for P. grandis and 52% for Q. quadrula (uncorrected amino acid p-distances). Phylogenetic analyses of 12 protein-coding genes from 29 bivalve and five outgroup mt genomes robustly indicate bivalve monophyly and the following branching order within the autolamellibranch bivalves: ((Pteriomorphia, Veneroida) Unionoida).
Conclusion: The basal nature of the Unionoida within the autolamellibranch bivalves and the previously hypothesized single origin of DUI suggest that (1) DUI arose in the ancestral autolamellibranch bivalve lineage and was subsequently lost in multiple descendant lineages and (2) the mitochondrial genome characteristics observed in unionoid bivalves could more closely resemble the DUI ancestral condition. Descriptions and comparisons presented in this paper are fundamental to a more complete understanding regarding the origins and consequences of DUI.
Figures

References
-
- Attardi G. Animal mitochondrial DNA: an extreme example of genetic economy. Int Rev Cytol. 1985;93:93–145. full_text. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

