Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces
- PMID: 20167085
- PMCID: PMC2838874
- DOI: 10.1186/1471-2180-10-51
Genetic adaptation of Streptococcus mutans during biofilm formation on different types of surfaces
Abstract
Background: Adhesion and successful colonization of bacteria onto solid surfaces play a key role in biofilm formation. The initial adhesion and the colonization of bacteria may differ between the various types of surfaces found in oral cavity. Therefore, it is conceivable that diverse biofilms are developed on those various surfaces. The aim of the study was to investigate the molecular modifications occurring during in vitro biofilm development of Streptococcus mutans UA159 on several different dental surfaces.
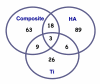
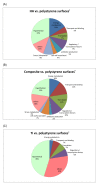
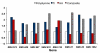
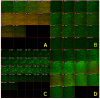
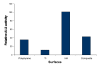
Results: Growth analysis of the immobilized bacterial populations generated on the different surfaces shows that the bacteria constructed a more confluent and thick biofilms on a hydroxyapatite surface compared to the other tested surfaces. Using DNA-microarray technology we identified the differentially expressed genes of S. mutans, reflecting the physiological state of biofilms formed on the different biomaterials tested. Eight selected genes were further analyzed by real time RT-PCR. To further determine the impact of the tested material surfaces on the physiology of the bacteria, we tested the secretion of AI-2 signal by S. mutans embedded on those biofilms. Comparative transcriptome analyses indicated on changes in the S. mutans genome in biofilms formed onto different types of surfaces and enabled us to identify genes most differentially expressed on those surfaces. In addition, the levels of autoinducer-2 in biofilms from the various tested surfaces were different.
Conclusions: Our results demonstrate that gene expression of S. mutans differs in biofilms formed on tested surfaces, which manifest the physiological state of bacteria influenced by the type of surface material they accumulate onto. Moreover, the stressful circumstances of adjustment to the surface may persist in the bacteria enhancing intercellular signaling and surface dependent biofilm formation.
Figures
Similar articles
-
DNA-microarrays identification of Streptococcus mutans genes associated with biofilm thickness.BMC Microbiol. 2008 Dec 29;8:236. doi: 10.1186/1471-2180-8-236. BMC Microbiol. 2008. PMID: 19114020 Free PMC article.
-
Streptococcus gordonii LuxS/autoinducer-2 quorum-sensing system modulates the dual-species biofilm formation with Streptococcus mutans.J Basic Microbiol. 2017 Jul;57(7):605-616. doi: 10.1002/jobm.201700010. Epub 2017 May 9. J Basic Microbiol. 2017. PMID: 28485524
-
Differential gene expression profiling of Streptococcus mutans cultured under biofilm and planktonic conditions.Microbiology (Reading). 2007 May;153(Pt 5):1307-1317. doi: 10.1099/mic.0.2006/002030-0. Microbiology (Reading). 2007. PMID: 17464045
-
Streptococcus mutans: a new Gram-positive paradigm?Microbiology (Reading). 2013 Mar;159(Pt 3):436-445. doi: 10.1099/mic.0.066134-0. Epub 2013 Feb 7. Microbiology (Reading). 2013. PMID: 23393147 Free PMC article. Review.
-
Communication among oral bacteria.Microbiol Mol Biol Rev. 2002 Sep;66(3):486-505, table of contents. doi: 10.1128/MMBR.66.3.486-505.2002. Microbiol Mol Biol Rev. 2002. PMID: 12209001 Free PMC article. Review.
Cited by
-
Bioadhesion in the oral cavity and approaches for biofilm management by surface modifications.Clin Oral Investig. 2020 Dec;24(12):4237-4260. doi: 10.1007/s00784-020-03646-1. Epub 2020 Oct 27. Clin Oral Investig. 2020. PMID: 33111157 Free PMC article. Review.
-
Decoding gene expression dynamics in planktonic and biofilm cells of Streptococcus mutans: regulation and role of mutanofactin genes in biofilm formation.Front Oral Health. 2025 Jan 17;6:1535034. doi: 10.3389/froh.2025.1535034. eCollection 2025. Front Oral Health. 2025. PMID: 39896144 Free PMC article.
-
CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans.PLoS One. 2013;8(3):e58271. doi: 10.1371/journal.pone.0058271. Epub 2013 Mar 12. PLoS One. 2013. PMID: 23554881 Free PMC article.
-
Exposure of Streptococcus mutans and Streptococcus sanguinis to blue light in an oral biofilm model.Lasers Med Sci. 2020 Apr;35(3):709-718. doi: 10.1007/s10103-019-02903-4. Epub 2019 Nov 12. Lasers Med Sci. 2020. PMID: 31713778
-
Effect of periodontal pathogens on the metatranscriptome of a healthy multispecies biofilm model.J Bacteriol. 2012 Apr;194(8):2082-95. doi: 10.1128/JB.06328-11. Epub 2012 Feb 10. J Bacteriol. 2012. PMID: 22328675 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

