Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference
- PMID: 20167086
- PMCID: PMC2843669
- DOI: 10.1186/1471-2407-10-48
Promoter methylation analysis of O6-methylguanine-DNA methyltransferase in glioblastoma: detection by locked nucleic acid based quantitative PCR using an imprinted gene (SNURF) as a reference
Abstract
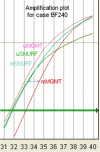
Background: Epigenetic silencing of the MGMT gene by promoter methylation is associated with loss of MGMT expression, diminished DNA-repair activity and longer overall survival in patients with glioblastoma who, in addition to radiotherapy, received alkylating chemotherapy with carmustine or temozolomide. We describe and validate a rapid methylation sensitive quantitative PCR assay (MS-qLNAPCR) using Locked Nucleic Acid (LNA) modified primers and an imprinted gene as a reference.
Methods: An analysis was made of a database of 159 GBM patients followed between April 2004 and October 2008. After bisulfite treatment, methylated and unmethylated CpGs were recognized by LNA primers and molecular beacon probes. The SNURF promoter of an imprinted gene mapped on 15q12, was used as a reference. This approach was used because imprinted genes have a balanced copy number of methylated and unmethylated alleles, and this feature allows an easy and a precise normalization.
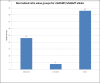
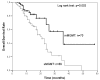
Results: Concordance between already described nested MS-PCR and MS-qLNAPCR was found in 158 of 159 samples (99.4%). The MS-qLNAPCR assay showed a PCR efficiency of 102% and a sensitivity of 0.01% for LNA modified primers, while unmodified primers revealed lower efficiency (69%) and lower sensitivity (0.1%). MGMT promoter was found to be methylated using MS-qLNAPCR in 70 patients (44.02%), and completely unmethylated in 89 samples (55.97%). Median overall survival was of 24 months, being 20 months and 36 months, in patients with MGMT unmethylated and methylated, respectively. Considering MGMT methylation data provided by MS-qLNAPCR as a binary variable, overall survival was different between patients with GBM samples harboring MGMT promoter unmethylated and other patients with any percentage of MGMT methylation (p = 0.003). This difference was retained using other cut off values for MGMT methylation rate (i.e. 10% and 20% of methylated allele), while the difference was lost when 50% of MGMT methylated allele was used as cut-off.
Conclusions: We report and clinically validate an accurate, robust, and cost effective MS-qLNAPCR protocol for the detection and quantification of methylated MGMT alleles in GBM samples. Using MS-qLNAPCR we demonstrate that even low levels of MGMT promoter methylation have to be taken into account to predict response to temozolomide-chemotherapy.
Figures


Similar articles
-
MGMT promoter methylation in patients with glioblastoma: is methylation-sensitive high-resolution melting superior to methylation-sensitive polymerase chain reaction assay?J Neurosurg. 2019 Mar 1;130(3):780-788. doi: 10.3171/2017.11.JNS171710. Epub 2018 May 4. J Neurosurg. 2019. PMID: 29726772
-
MGMT gene silencing and benefit from temozolomide in glioblastoma.N Engl J Med. 2005 Mar 10;352(10):997-1003. doi: 10.1056/NEJMoa043331. N Engl J Med. 2005. PMID: 15758010
-
MGMT promoter methylation correlates with survival benefit and sensitivity to temozolomide in pediatric glioblastoma.Pediatr Blood Cancer. 2007 Apr;48(4):403-7. doi: 10.1002/pbc.20803. Pediatr Blood Cancer. 2007. PMID: 16609952
-
Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity.J Clin Oncol. 2008 Sep 1;26(25):4189-99. doi: 10.1200/JCO.2007.11.5964. J Clin Oncol. 2008. PMID: 18757334 Review.
-
O6-methylguanine DNA methyltransferase gene promoter methylation status in gliomas and its correlation with other molecular alterations: first Indian report with review of challenges for use in customized treatment.Neurosurgery. 2010 Dec;67(6):1681-91. doi: 10.1227/NEU.0b013e3181f743f5. Neurosurgery. 2010. PMID: 21107199 Review.
Cited by
-
The prognostic value of MGMT promoter methylation in Glioblastoma multiforme: a meta-analysis.Fam Cancer. 2013 Sep;12(3):449-58. doi: 10.1007/s10689-013-9607-1. Fam Cancer. 2013. PMID: 23397067
-
DNA Methylation Description of Hippocampus, Cortex, Amygdala, and Blood of Drug-Resistant Temporal Lobe Epilepsy.Mol Neurobiol. 2023 Apr;60(4):2070-2085. doi: 10.1007/s12035-022-03180-z. Epub 2023 Jan 5. Mol Neurobiol. 2023. PMID: 36602701
-
Unveiling the role of O(6)-methylguanine-DNA methyltransferase in cancer therapy: insights into alkylators, pharmacogenomics, and others.Front Oncol. 2024 Jul 11;14:1424797. doi: 10.3389/fonc.2024.1424797. eCollection 2024. Front Oncol. 2024. PMID: 39055560 Free PMC article. Review.
-
Development of a multiplex methylation-specific PCR as candidate triage test for women with an HPV-positive cervical scrape.BMC Cancer. 2012 Nov 23;12:551. doi: 10.1186/1471-2407-12-551. BMC Cancer. 2012. PMID: 23176198 Free PMC article.
-
Correlation of MLH1 and MGMT methylation levels between peripheral blood leukocytes and colorectal tissue DNA samples in colorectal cancer patients.Oncol Lett. 2013 Nov;6(5):1370-1376. doi: 10.3892/ol.2013.1543. Epub 2013 Aug 23. Oncol Lett. 2013. PMID: 24179526 Free PMC article.
References
-
- Komine C, Watanabe T, Katayama Y, Yoshino A, Yokoyama T, Fukushima T. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13(2):176–184. - PMC - PubMed
-
- Qian XC, Brent TP. Methylation hot spots in the 5' flanking region denote silencing of the O6-methylguanine-DNA methyltransferase gene. Cancer Res. 1997;57(17):3672–3677. - PubMed
-
- Watts GS, Pieper RO, Costello JF, Peng YM, Dalton WS, Futscher BW. Methylation of discrete regions of the O6-methylguanine DNA methyltransferase (MGMT) CpG island is associated with heterochromatinization of the MGMT transcription start site and silencing of the gene. Mol Cell Biol. 1997;17(9):5612–5619. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

